Scientists reveal the molecular basis of blocking host xenophagy by Salmonella effector SopF and common mechanism underlying diverse types of selective autophagy
Autophagy used to be considered as a nonselective bulk degradation pathway that delivers cellular constituents into lysosomes to help cells cope with nutrient deprivation. Recent studies demonstrated that autophagic machinery can also selectively recognize specialized cargos including invading bacteria and other pathogens and initiate bacterial autophagy (xenophagy) that antagonizes bacterial proliferation in host cells. The underlying mechanism of xenophagy was first discovered by Prof. SHAO Feng's group (National Institute of Biological Sciences) in 2019. Invading bacteria residing in endosome-like vacuoles cause membrane damages to the vacuoles owing to their pore-forming toxins or secretion apparatus. The V-ATPase, abundantly present on the bacteria-containing vacuoles, perceives the damages and recruits the ATG16L1 complex that then signals the ATG machinery to induce LC3 lipidation. LC3-decorated compartments then undergo lysosome-mediated degradation. In this work, a Salmonella T3SS effector SopF was also identified as a specific inhibitor for host xenophagy. SopF induces site-specific ADP-ribosylation of the VoC subunit of V-ATPase complex, which prevents the recruitment of ATG16L1 and thereby blocks formation of the LC3-marked autophagosome. However, the precise molecular basis of SopF inhibition of xenophagy, as well as the possible mechanism of xenophagy initiated by V-ATPase-ATG16L1 axis remains to be fully explored.
On January 19, 2022, Nature Structural & Molecular Biology published an article entitled "ARF GTPases activate Salmonella effector SopF to ADP-ribosylate host V-ATPase and inhibit endomembrane damage-induced autophagy" by a collaborative study of Prof. WANG Dacheng/DING Jingjin's group (Institute of Biophysics, Chinese Academy of Sciences) and Prof. SHAO Feng's group (National Institute of Biological Sciences). This work revealed that SopF employs GTP-bound host small GTPase ARF as a cofactor to catalyze the ADP-ribosylation of VoC subunit of V-ATPase and thereby blocks xenophagy. Moreover, they discovered that activation of V-ATPase-ATG16L1 pathway is universally involved in LC3 activation upon damage of various endomembranes.
By mass-spectrometry analyses of SopF immunoprecipitates from 293T cells, researchers identified host small GTPase ARF proteins that specifically bind to SopF. Furthermore, they determined the crystal structures of NAD+-free and NAD+-bound SopF-ARF1 complexes. Structural and mutagenesis analyses revealed that SopF is a diphtheria toxin-like bona fide ADP-ribosyltransferase. SopF adopts an "I-Y-E" motif of catalytic triad to bind NAD+ donor to catalyze ADP-ribosylation of Gln124 in the VoC subunit of V-ATPase complex. Bacterial infection and in vitro biochemical results suggested that binding with GTP-bound ARF is crucial for SopF ADP-ribosyltransferase activity. Employing host proteins to activate functional activities of bacterial effectors implicates a sophisticated strategy evolved by long-term bacteria-host interactions.
Given that V-ATPase is a widely distributed proton pump on endomembrane organelles, SopF is supposed to be a powerful tool to investigate whether V-ATPase-ATG16L1 axis also play a critical role in other types of selective autophagy caused by endomembrane damages. Following this hypothesis, they found that LLOMe, Nigericin or NH4Cl treatment can induce activation on damaged lysosomes, which can be blocked by SopF or Q124A mutation of VoC, suggesting that selective autophagy in response to lysosomal damage is likely initiated by V-ATPase-ATG16L1 axis. Recent studies showed that STING, a key protein in interferon-mediated antiviral pathway, upon binding to its ligand cGAMP oligomerizes and translocates to Golgi apparatus. This process induces LC3 activation on Golgi apparatus. Consistently, ectopic expression of SopF or introduction of VoC Q124A mutation both inhibited LC3 decoration on Golgi apparatus, implying selective autophagy in response to Golgi perturbation also relies on the V-ATPase-ATG16L1 axis. More interestingly, they found that SopF or Q124A mutation of VoC subunit did not impair the proton-pumping activity of V-ATPase. V-ATPase acts as a sensor of pH change caused by endomembrane damages, which then recruits ATG16L1 to initiate selective autophagy.
This study revealed the structural basis for the ADP-ribosyltransferase activity of bacterial effector SopF, as well as ARF-dependent regulation of SopF activity. Furthermore, they identified a novel function of V-ATPase to perceive endomembrane damages and initiate selective autophagy, which is independent of its proton-pump activity. More importantly, they established the V-ATPase-ATF16L1 axis as a common mechanism for diverse types of selective autophagy, providing a new direction for studying the process and regulation of endomembrane homeostasis.
Prof. DING Jingjin from Institute of Biophysics, Chinese Academy of Sciences and Prof. SHAO Feng from National Institute of Biological Sciences are the co-corresponding authors of this paper. This research was supported by the National Key Research and Development Program of China, the Strategic Priority Research Program of the Chinese Academy of Sciences, Excellent Young Scholar Program of the National Natural Science Foundation of China, and a grant from the CAS Youth Innovation Promotion Association.
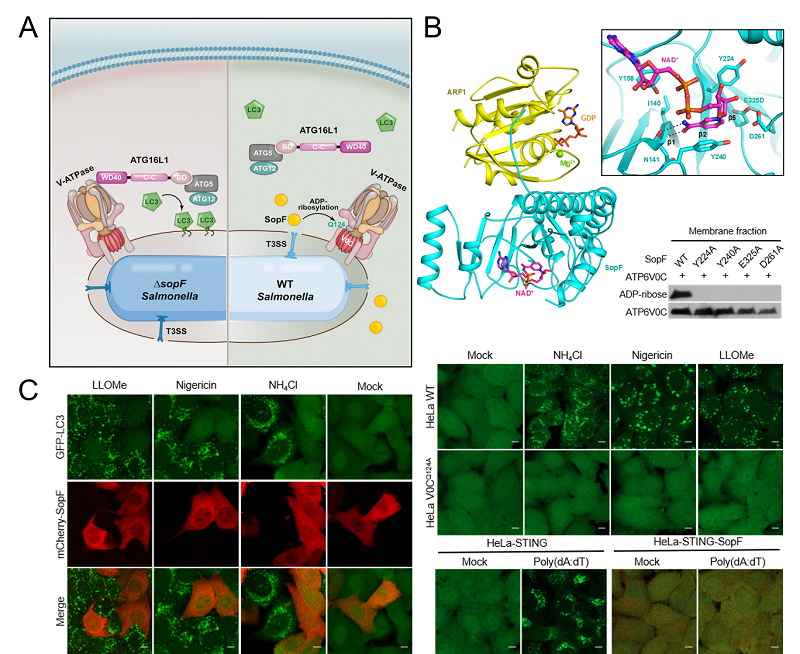
Figure. The molecular mechanism of blocking host xenophagy by Salmonella effector SopF
A: Schematic model of Salmonella effector SopF inhibition of xenophagy through blocking V-ATPase-ATG16L1 axis; B: Crystal structure of SopF-ARF1-NAD+ complex with close-up view of NAD+ binding pocket; C: Ectopic expression of SopF in HeLa cells inhibits diverse types of selective autophagy caused by endomembrane damages.
Article link:https://www.nature.com/articles/s41594-021-00710-6
Contact: DING Jingjin
Institute of Biophysics, Chinese Academy of Sciences
Beijing 100101, China
Email: jding@ibp.ac.cn
(Reported by Dr. WANG Dacheng/DING Jingjin's group)

