Review: Oxidative protein folding fidelity and redoxtasis in the endoplasmic reticulum
Over one third of human protein-coding genes are predicted to encode secretory or membrane proteins. These proteins are enriched with disulfide bonds, which are important to their structure, function and regulation. Disulfide bonds form during nascent peptide folding into native proteins in a process called oxidative protein folding, which takes place mainly in the lumen of the endoplasmic reticulum (ER) in eukaryotic cells. The environment of the ER is optimized for oxidative protein folding. The ER contains a variety of folding enzymes and molecular chaperones. In addition, the redox state of the ER is more oxidizing than that in the cytosol, in terms of the main cellular redox buffer, reduced/oxidized glutathione (GSH/GSSG). The glutathione reduction potential (EGSH) in the ER is approximately -200 mV, which is much higher than the value of approximately -300 mV in the cytosol. Nevertheless, perturbation of ER redox homeostasis (redoxtasis) and accumulation of unfolded/misfolded proteins can lead to ER stress and related diseases. Therefore, studies aimed at determining the mechanism through which cells guarantee the efficiency and fidelity of oxidative protein folding and maintain ER redoxtasis are important.
On 20th July, 2022, Trends in Biochemical Science published an invited review paper entitled "Oxidative protein folding fidelity and redoxtasis in the endoplasmic reticulum", written by Prof. WANG Lei and Prof. WANG Chih-chen at the Institute of Biophysics, Chinese Academy of Science (CAS). In this review, the authors summarized oxidative protein folding machineries in yeast, plants, and mammals and focused on the mechanisms how the oxidative protein folding fidelity and ER redoxtasis are maintained. In addition, they discussed the role of oxidative protein folding fidelity in different pathophysiology and the possibility of targeting oxidative protein folding for disease interventions.
The ER sulfhydryl oxidase 1 (Ero1) and protein disulfide isomerase (PDI) family constitute a pivotal pathway for oxidative protein folding in the ER. In this review, the authors proposed that oxidative protein folding fidelity and ER redox homeostasis are maintained by two key mechanism: 1) Ero1 oxidase determines the relatively high oxidizing environment of the ER, and its activity is precisely controlled by various posttranslational modifications (PTMs) on its 'regulatory loop'; 2) The abundant PDI family members in higher eukaryotes exhibit division of labor during oxidative protein folding, and they work synergistically to guarantee efficient and faithful disulfide formation. In the future, a better understanding of the regulatory mechanism of oxidative protein folding and the discovery of specific inhibitors of Ero1-PDI interplay will certainly facilitate the development of novel strategies for the treatment of related diseases.
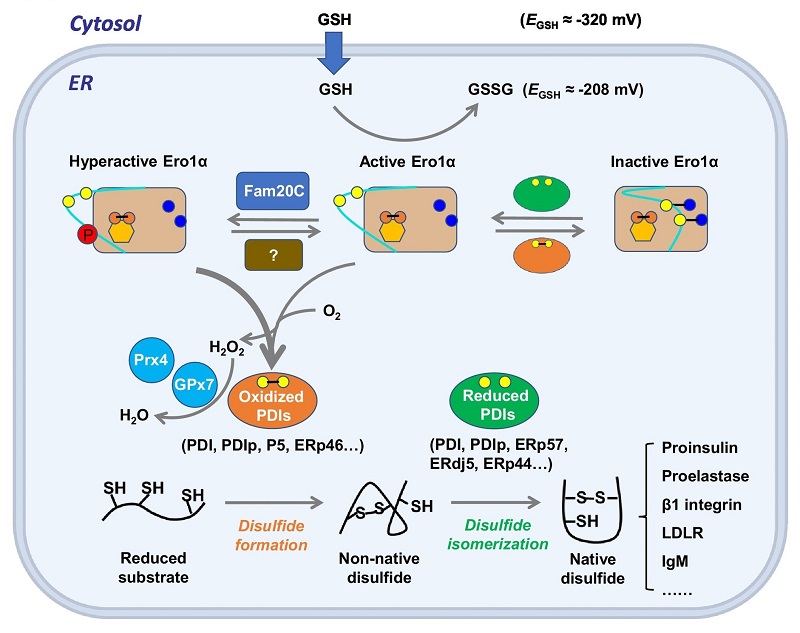
The Ero1α-PDI system safeguards oxidative protein folding fidelity and redoxtasis in the ER.
Article link:
https://www.cell.com/trends/biochemical-sciences/fulltext/S0968-0004(22)00168-2
Contact: WANG Lei
Institute of Biophysics, Chinese Academy of Sciences
Beijing 100101, China
Email: wanglei@ibp.ac.cn
(Reported by Dr. WANG Chih-chen/Wang Lei's group)

