Researchers Reveal the Transcriptional Mechanism of Mammalian Reovirus
Mammalian reovirus (MRV), a member of the Reoviridae family, has three major serotypes: type 1 Lang (T1L), type 2 Jones (T2J), and type 3 Dearing (T3D). Specifically, T3D preferentially induces cell death and apoptosis in tumor cells but not in nontransformed cells making it a potential oncolytic agent. A recent study revealed that the most oncolytic T3D strain transcribes at rates superior to those of the less oncolytic T3D strains. Thus, understanding the molecular mechanism of transcription stimulation by uncoating the outer layer capsid and transcription regulation will be conducive to the development of reovirus-based therapies with heightened antitumor efficiency.
MRV is the prototypical member of genus Orthoreovirus of family Reoviridae. It has a 10-segment dsRNA genome surrounded by two icosahedral protein layers. During cell entry, outer capsid is proteolytically degraded remaining the transcriptionally active core particle. In the core, the RNA-dependent RNA polymerase (RdRp, λ3 protein), which is complexed with cofactor μ2 and located beneath the five-fold axis of the core shell, uses the negative strand of each genomic dsRNA segment as a template to synthesize nascent mRNA. After transcription, the nascent transcript is capped by turret protein λ2, which is further extruded into the cytosol and translated into viral proteins.
Recently, a research group led by Prof. ZHU Ping at the Institute of Biophysics, Chinese Academy of Sciences (CAS), and their collaborators describe the in situ structures of the reovirus transcriptase complex in the intact double-layered virion, and the single-layered uncoated cores in the unloaded state, reloaded state, pre-elongation state, and elongation state, respectively, by cryo-EM and sub-particle reconstruction. This study has been published online ahead of print as a research article entitled “In situ Structures of Polymerase Complex of Mammalian Reovirus Illuminate RdRp Activation and Transcription Regulation” in the Proceedings of the National Academy of Sciences (PNAS) on Dec. 5, 2022.
The newly published study revealed the structural details of variant viral proteins, particularly the RdRp and associated proteins, during transcription. In virion, the genomic dsRNA is densely packaged inside the core, while the terminal genomic RNA is unwound by a positively charged patch formed by RdRp and μ2 at the template entrance to form a transcription initiation state. After uncoating of the outer capsid shell, the RNA-loading region of RdRp near the template entry channel becomes flexible making the RNAs unloaded. Upon adding essential materials (SAM/ATP/UTP/CTP), RdRp transfers to a reloaded state that is characterized mainly by the recovery of the RNA-loading region and the genomic terminal RNAs in the core. RdRp is then further transferred to the pre-elongation state, where N-MTase domain is ligand-bound. Once total transcriptional substrates added, RdRp is thoroughly enabled into the elongation state. With the growing of the transcript, the priming loop is pushed away from the trajectory of the template-transcript duplex. The rearranged priming loop in turn pushes the latch helix away, which further leads to conformational rearrangement of the RdRp C-terminal domain (CTD). Consequently, the transcript exit is exposed by the transition of the loop1099-1118 from a latch helix configuration to a latch loop, and the wedge helix shifts to the right position of the template-transcript hybrid, making the conserved residue Asp1120 rightly insert and split the duplex. Once separated, the template strand exits through the template exit channel and reanneals with the encoding chain, which is achieved by the cooperation of positively charged regions in the NTD of μ2 and CTD of RdRp. At the same time, the nascent transcript successively passes through the mRNA exit channel of RdRp and then the core shell exit pore right at the five-fold vertex of the capsid for subsequent capping by the turret protein λ2.
This study shed light on the assembly mechanism of transcriptase complex into reovirus virion. With the integration of these structures, a transcriptional model of reovirus with five states is proposed. The structures illuminate the RdRp activation and regulation of the multilayered turreted reovirus, and may contribute to the development of reovirus-based therapies.
This work was supported by grants from the Ministry of Science and Technology of China, the National Natural Science Foundation of China and the Chinese Academy of Sciences.
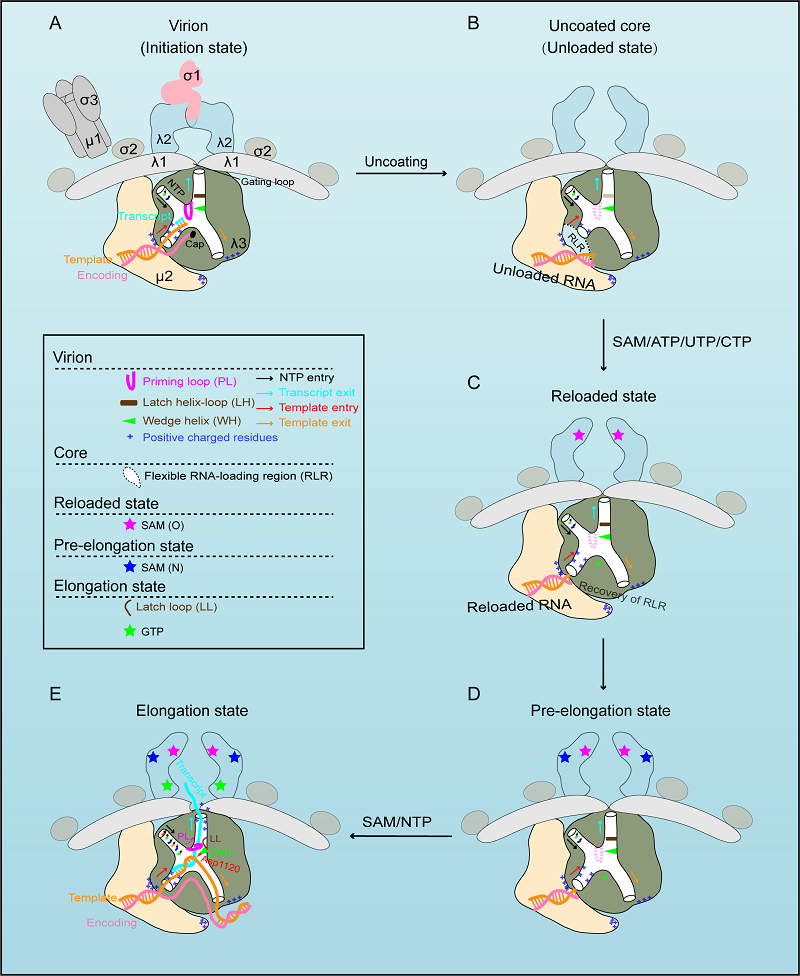
A transcriptional model of Mammalian reovirus
(Image by Dr. ZHU Ping’s group)
The study can be found online ahead of publication of PNAS:
https://www.pnas.org/doi/10.1073/pnas.2203054119
Contact: ZHU Ping
Institute of Biophysics, Chinese Academy of Sciences
Beijing 100101, China
Email: zhup@ibp.ac.cn
(Reported by Dr. ZHU Ping's group)

