Heterogeneity of ribosomes: Different ribosomes produce different proteomes
Background:
Ribosomes are one of the most important polymerases that synthesize proteins using messenger RNA (mRNA) as a template and aminoacylated tRNA (aa-tRNA) as a substrate. Ribosomes have millions to tens of millions of copies in each mammalian cell (1, 2). ribosomal RNA (rRNA) accounted for 80-85% of the total RNA in all cell types, and ribosomal protein (rProtein) accounted for 5-10% of the total protein volume in mammals. 10-20% of the total protein copy number (1).
Ribosomes are direct spheroids at 20 to 30nm, and a wide range of ribosomes can be seen in high resolution imaging of cells. In rapidly proliferating cells, ribosomes occupy a higher proportion of matter and energy, organizing cell genesis and development and determining cell fate (Qin Yan's research group reviewed in the Science Bulletin, July 2022 (3)). Special ribosomes can produce special proteome, so as to determine the function and fate of cells, which is an important proposition in recent years. The production of proteins, the main carrier of cell function, is a problem to trace back to the root: how the ribosome determines the proteome.
The article found that
In male germ cells, the ribosomal large subunit protein L39 is transferred after spermatogenic meiosis and uses the form L39L on the ribosome. Our study found that L39 is an important component of the neopeptide channel of the large subunit of the ribosome. After L39L replaces L39, this channel can be much wider, which is conducive to the production of a large number of positive proteins during sperm maturation.
In this study, how does ribosome L39 determine the sperm germ proteome? The mechanism was studied mainly through the superresolved cryo-EM structure analysis of L39L-type RibosomeST and ordinary L39L-type Ribosome (the resolution is 2.82 angstroms in L39 and 3.03 angstroms in L39L). We found the important components of the large subunit neopeptide channel of ribosome. After L39L replaces L39, this channel can be much wider, which is conducive to the production of a large number of positive proteins during sperm maturation (Figure 1).
Spermatogenesis is the core function of species reproduction. Oligasthenospermia has become an important national health problem in recent years. The study of L39 provides an important marker and therapeutic target for this disease.
Referee: It reflects a ground-breaking story that is exceptional and essential for our knowledge of ribosomes in organismal development and control of gene expression. The study is beautiful as it combines mouse genetics, detailed gene expression studies, state of the art proteomics, detailed biochemical insight, and CryoEM.
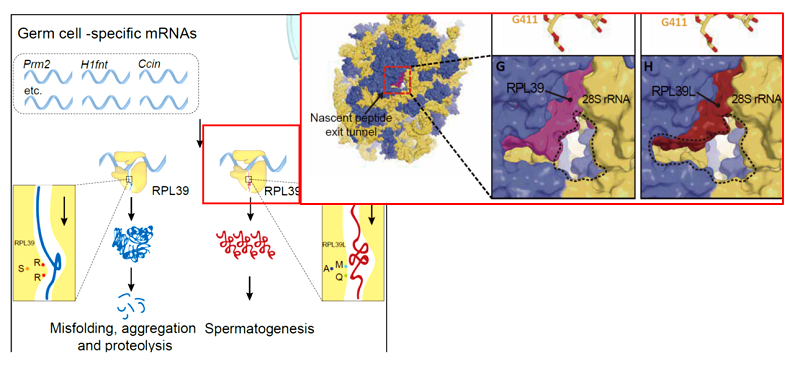
Fig. 1 The special ribosome-ribosomest in spermatogenesis, where L39L (right, red protein) replaces the normal L39 (right, purple protein), results in a widening of the ribosomal neopeptide channel, contributing to the production of a large number of positively charged neopeptide chains in spermatogenesis(Li et al., Nature, 2022)。
In that perspective, while the studies largely overlap (and thus provide convincing support of the specific ribosome/translation-linked role of RPL39L), the current study provides further insights in the possible molecular mechanism of action in that CryoEM structural analysis were conducted demonstrating distinct features of the ribosomal polypeptide exit tunnel (charge state and size) of the specialized ribosome, possibly implicating these distinct features in the folding of a subset of male germ cell specific proteins, besides demonstrating the importance of Q28, as well as the noninterchangeable ribosomal functions of RPL39L and RPL39.
Referee: The authors addressed all my comments, and this fascinating paper is ready for publication. I believe that this work represents a breakthrough in our understanding of how specialized ribosomes affect gene expression and organismal development.
This study was funded by the National Key R&D Program of China and the National Natural Science Foundation of China.
Article link: https://www.nature.com/articles/s41586-022-05508-0
Reference
1. An, H., and Harper, J. W. (2020) Ribosome Abundance Control Via the Ubiquitin-Proteasome System and Autophagy. J Mol Biol 432, 170-184
2. An, H., Ordureau, A., Korner, M., Paulo, J. A., and Harper, J. W. (2020) Systematic quantitative analysis of ribosome inventory during nutrient stress. Nature 583, 303-309
3. Wang, Q., Wang, Y. B., Li, S. G., Zhou, A. Q., and Qin, Y. (2022) Organelle biogenesis: ribosomes as organizer and performer. Science Bulletin 67, 1614-1617
4. Liu, G., Song, G., Zhang, D., Zhang, D., Li, Z., Lyu, Z., Dong, J., Achenbach, J., Gong, W., Zhao, X. S., Nierhaus, K. H., and Qin, Y. (2014) EF-G catalyzes tRNA translocation by disrupting interactions between decoding center and codon-anticodon duplex. Nat Struct Mol Biol 21, 817-824
5. Zhang, D., Yan, K., Liu, G., Song, G., Luo, J., Shi, Y., Cheng, E., Wu, S., Jiang, T., Lou, J., Gao, N., and Qin, Y. (2016) EF4 disengages the peptidyl-tRNA CCA end and facilitates back-translocation on the 70S ribosome. Nat Struct Mol Biol 23, 125-131
Contact: QIN Yan
Institute of Biophysics, Chinese Academy of Sciences
Beijing 100101, China
Email: qiny@ibp.ac.cn
(Reported by Dr. QIN Yan's group)

