Scientists reveal the molecular mechanisms of calmodulin activating Shigella effector OspC3 to ADP-riboxanate caspase-4/11 and block pyroptosis
Pyroptosis is one of the most essential innate immune defenses against infection by pathogens, especially bacterial invaders. When Gram-negative bacteria get into host cells, their conserved outer membrane component lipopolysaccharide (LPS), also regarded as a well-known pathogen-associated molecular pattern (PAMP), could be recognized by host intracellular innate immune receptors caspase-4/5/11. LPS-activated caspase-4/5/11 cleave gasdermin D (GSDMD) to unmask the pore-forming activity of GSDMD, which lyses host cells to trigger pyroptosis and antibacterial immune responses. On the other hand, bacteria evolve versatile strategies to evade host immune defenses. For instance, they often "inject" various virulence effectors into host cells through specialized Type III secretion system (T3SS) to hijack host innate immune pathways. In 2021, Prof. SHAO Feng's group (National Institute of Biological Sciences, Beijing) discovered that Shigella fexneri employs a T3SS effector OspC3 to exclusively block host caspase-4/11-GSDMD pathway. OpsC3 catalyzes an unprecedented posttranslational modification (PTM) called ADP-riboxanation on a crucial arginine residue located in the active center of caspase-4/11, and inactivates these caspases to block the cleavage of GSDMD and the resulting pyroptosis. However, how Shigella OspC3 specifically targets host caspase-4/11 and catalyzes the novel PTM ADP-riboxanation remain unknown.
On Jan 9, 2023, the journal Nature Structural & Molecular Biology published online an article entitled "Structural mechanisms of calmodulin activation of Shigella effector OspC3 to ADP-riboxanate caspase-4/11 and block pyroptosis" by a collaborative study of Prof. WANG Dacheng/DING Jingjin's group (Institute of Biophysics, Chinese Academy of Sciences) and Prof. SHAO Feng's group (National Institute of Biological Sciences, Beijing). This work establishes a comprehensive framework for understanding the molecular mechanisms by which OspC3 employs host Ca2+-free calmodulin (CaM) protein as a cofactor to catalyze the novel ADP-riboxanation of caspase-4/11 and block host caspase-4/11-GSDMD pyroptosis pathway.
OspC3 was found to effectively modify both inactive and autoprocessed active forms of caspase-4/11. However, when the enzymatic active center of caspase-4/11 was irreversibly occupied by a substrate-mimicking inhibitor zVAD, OspC3 modification on these zVAD-bound caspases was largely impaired. This observation indicates that OspC3 prefers the substrate-free forms of caspase-4/11, whether activated or not. The researchers further characterized that ADP-ribose-binding macro-domain protein Af1521 can bind ADP-riboxanated caspase-4/11 to form a 1:1 stable complex. They determined the 1.96-Å crystal structure of ADP-riboxanated caspase-4-Af1521 complex which illustrates for the first time the precise chemical structure of ADP-riboxanation. A new five-atom oxazolidine ring is formed by covalent attachment of ADP-ribose onto the deaminated guanidine group of caspase-4 R314, in which the C1 and 2'-OH of the ribose group in ADP-ribose are linked to the Nδ and C atoms of the deaminated guanidine group of R314, respectively. This crystal structure provides a solid structural evidence for the unprecedented PTM of arginine ADP-riboxanation.
OspC3 belongs to the OspC family of bacterial effectors which adopt a conserved two-domain architecture. The N-terminal catalytic domain shares no significant sequence homologies with any known proteins, whereas the C-terminal domain comprises a typical ankyrin-repeat domain (ARD) that often mediates protein-protein interactions. The researchers solved the crystal structure of OspC3ARD-caspase-4 complex and identified the detailed hydrogen-bond network and hydrophobic interactions responsible for OspC3 recruitment of host caspase-4/11 substrates via the C-terminal ARD.
In an in vitro biochemical reconstitution system to capture OspC3 modification of caspase-4/11, the researchers found that equal molar amount of enzyme was required for complete ADP-riboxanation of the substrate. This is contrary to the well-established knowledge about the high efficiency of an enzyme-catalyzing reaction. Mass-spectrometry analyses of OspC3 immunoprecipitates from Hela cells identified calmodulin (CaM) as a potential binder of OspC3. OspC3 selectively binds Ca2+-free CaM to form a 1:1 stable complex. Addition of Ca2+-free CaM into the ADP-riboxanation reaction greatly potentiated OspC3 enzymatic activity as one-percent molar amount of OspC3 rendered complete modification of caspase-4. By solving the high-quality crystal structure of OspC3-CaM complex, the researchers revealed that CaM employs its two lobes to grasp the N-terminal domain of OspC3 through extensive hydrophobic interactions. The OspC3-N domain bears a typical Rossmann fold and shares the conserved NAD+-binding site with other classical ADP-ribosyltransferase (ART) domains. To fully understand the enzymatic mechanisms of OspC3 ADP-riboxanating caspase-4/11, the researchers further determined the crystal structures of OspC3-CaM-caspase-4 ternary complex and the quaternary complex with 2'-F-NAD+ (a nonhydrolyzable NAD+ analog). The complex structures show that the acidic residue D231 located in the active center of OspC3-N domain immobilizes the terminal Nω of caspase-4 R314, inducing a crooked conformation of the guanidine and exposing its Nδ for attacking C1 of the ADP-ribose derived from the scission of the glycosidic bond in NAD+ donor. This explains why the ADP-ribosylation in OspC3-mediated catalysis occurs on the Nδ rather than the Nω seen with canonical arginine ARTs. Another acidic residue D177 around the active center of OspC3-N domain serves as a base to activate the 2′-OH of ADP-ribose to attack the guanidine carbon of caspase-4 R314 for deamination, thereby completing the catalysis of ADP-riboxanation by forming an oxazolidine. Mechanisms derived from the structural findings are further supported by mutagenesis-based biochemical analyses in vitro and functional validation in S. flexneri-infected mice.
Through a series of structures covering the entire process of OspC3 action and extensive functional validations, this study uncovers the complete molecular mechanisms of Shigella effector OspC3 ADP-riboxanating innate immune receptor caspase-4/11 with the help of host cofactor CaM and blocking caspase-4/11-GSDMD pyroptosis pathway to antagonize host defense. It also provides a comprehensive understanding of the novel PTM ADP-riboxanation and offers a valuable strategy for developing new antibacterial drugs and bacterial live-attenuated vaccines.
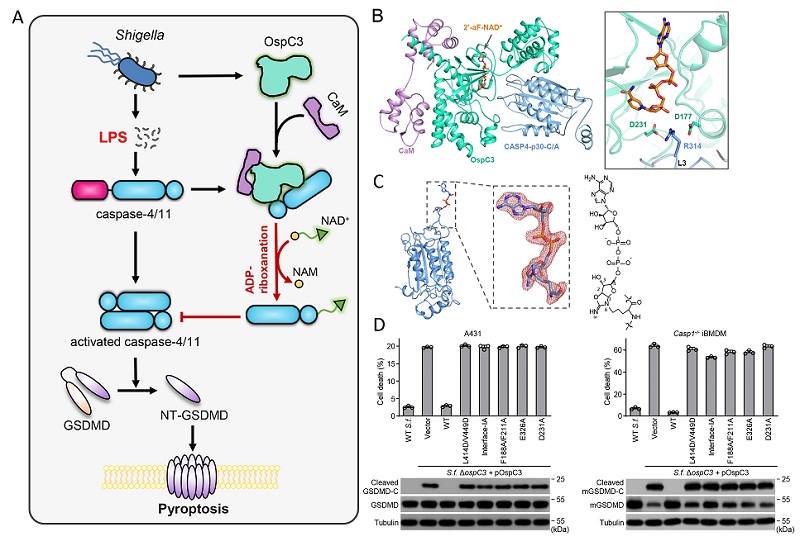
Figure. The molecular mechanism of Shigella effector OspC3 blocking host pyroptosis
A. Schematic model of Shigella effector OspC3 blocking host caspase-4/11-GSDMD pyroptosis pathway; B. Crystal structure of OspC3-CaM-caspase-4-2'-F-NAD+ quaternary complex in cartoons and the close-up view of OspC3 catalytic center; C. Structure of OspC3-modified caspase-4 with arginine ADP-riboxanation highlighted; D. The residues crucial for OspC3 ADP-riboxanating caspase-4/11 were validated by pyroptosis inhibition in S. flexneri-infected cells.
Prof. DING Jingjin from Institute of Biophysics, Chinese Academy of Sciences and Prof. SHAO Feng from National Institute of Biological Sciences are the co-corresponding authors of this work. Dr. HOU Yanjie from Ding Jingjin's group and PhD student ZENG Huan from SHAO Feng's group are the co-first authors of this paper. This research was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences, the National Key Research and Development Program of China, Excellent Young Scholar Program of the National Natural Science Foundation of China, and a grant from the CAS Youth Innovation Promotion Association.
Article link: https://www.nature.com/articles/s41594-022-00888-3
Contact: DING Jingjin
Institute of Biophysics, Chinese Academy of Sciences
Beijing 100101, China
Email: jding@ibp.ac.cn
(Reported by Dr. DING Jingjin's group)

