Structure of the enigmatic chloroplast protein transport machinery elucidated
Chloroplasts of algae and plants are the cellular engines that convert solar energy into chemical energy through photosynthesis. These organelles, bounded by an envelope with two membranes, contain their own genome whose expression is tightly coordinated with that of the nuclear genome (harboring most of the genetic information of the cells). The majority of chloroplast proteins, nearly 3000, are encoded by nuclear genes, translated in the cytosol as precursor proteins containing a transit sequence at their amino terminus that serves as the entry ticket into chloroplasts. Protein import into chloroplasts is mediated by two membrane protein complexes called TOC and TIC in the outer and inner envelope membrane, respectively. These complexes play a key role in chloroplast biogenesis, in the assembly of the photosynthetic apparatuses and in various metabolic pathways.
During the past 30 years, the different protein subunits of TOC and TIC have been identified and characterized. More recent work has revealed that TOC and TIC form a supercomplex together. However, it is unclear how the different proteins of TOC and TIC assemble together to form the channels for protein translocation across the chloroplast envelope membranes and the protein translocation pathways within TOC and TIC remain elusive.
On Jan. 26, LIU Zhenfeng’s group at the Institute of Biophysics, in collaboration with Prof. Jean-David Rochaix from the University of Geneva, Switzerland, published online a research work entitled “Architecture of chloroplast TOC-TIC translocon supercomplex” as an article in the journal Nature.

Figure 1: The TOC-TIC translocon supercomplex is responsible for the import of the prepoteins synthesized in the cytosol into chloroplasts.
In this work, the supramolecular architecture of the TOC-TIC supercomplex was elucidated through cryo-electron microscopy (Figure 1). Thirteen different protein subunits were discovered in this supercomplex. With exception of Tic214 encoded by the chloroplast genome, all the other subunits are nuclear encoded. They are assembled into the TOC complex in the outer membrane, the intermembrane space complex (ISC) and the TIC complex in the inner membrane. Remarkably, it was found that the largest membrane protein Tic214 spans the inner membrane, the intermembrane space and the outer membrane, linking the other protein subunits like a bridge and most likely also acting as a scaffold (Figure 2). The TOC complex on the outer membrane is mainly comprised of Toc34, Toc90 and Toc75, flanked on the Toc90 side by the Ctap4-Ctap3 complex. A hybrid barrel-shaped channel is formed by Toc90 and Toc75 on the outer membrane. The channel contains an entrance on the cytosolic side and two exits opening toward the intermembrane space, as well as a lateral gate facing the lipid bilayer. A phytic acid (also known as inositol hexaphosphate/InsP6) molecule intercalates at the interface between Toc90 and Tic214, stabilizing their assembly like a wedge. The intermembrane-space domain of Tic214, Tic100, Tic56, Ctap3 and Ctap5 intertwine with each other to form a tower-like structure connecting TOC with TIC. In the inner membrane, the membrane-embedded domains of Tic214, Tic20, Ctap5 and three small subunits (named Simp1, Simp2 and Simp3) form the TIC complex. Four lipid molecules serve to stabilize the assembly of a funnel-like channel located at the interface between Tic214 and Tic20 and prevent the channel from leaking.
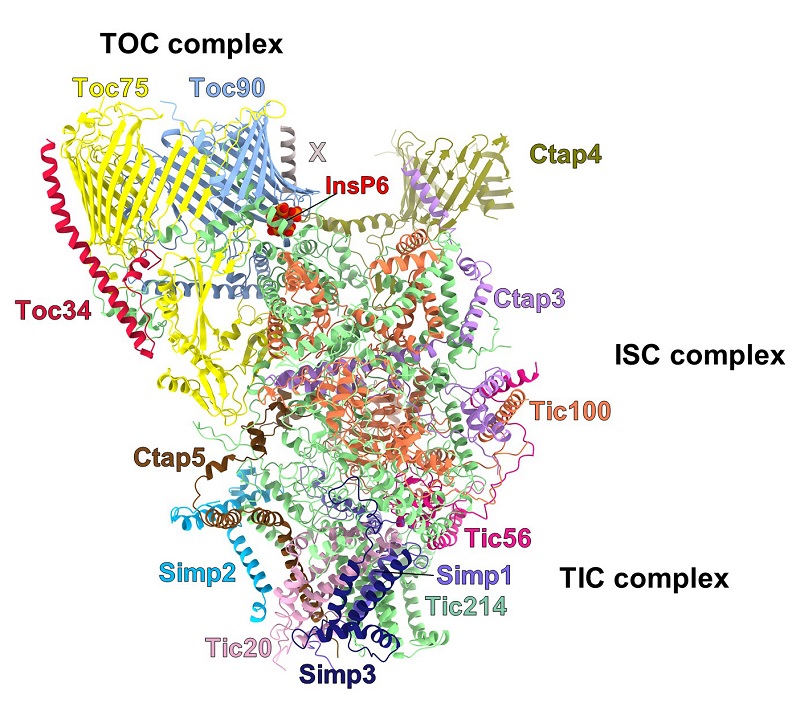
Figure 2:Subunit composition and assembly of the TOC-TIC supercomplex.
Based on the structural data, the researchers analyzed in detail the features of the pores inside the TOC and TIC channels. They were able to predict the interactions between the transit peptide and the TIC complex through molecular dynamics simulation. Supported by the observation of two channel pores connected by a surface groove and the previous biochemical and functional reports, multiple pathways were proposed to account for the translocation of the different preproteins through the TOC-TIC supercomplex to distinct local chloroplast compartments.
LIU Hao and LI Anjie, the graduate students from the Institute of Biophysics, Chinese Academy of Sciences, are the first and second author of the paper respectively. Prof. Jean-David Rochaix (from the University of Geneva, Switzerland) participated in the project design, analysis of the results and discussion. Dr. LIU Zhenfeng is the corresponding author of the work. The project was funded by the National Natural Science Foundation of China, Chinese Academy of Sciences and the Chinese Ministry of Science and Technology.
(Credit: LIU Hao, Jean-David Rochaix and LIU Zhenfeng)
Link to the paper:https://www.nature.com/articles/s41586-023-05744-y
Contact: LIU Zhenfeng
Institute of Biophysics, Chinese Academy of Sciences
Beijing 100101, China
Email: liuzf@ibp.ac.cn
(Reported by Dr. LIU Zhenfeng's group)

