Researchers develop a CRIC-seq method to decipher positional rules of PTBP1-associated RNA loops in regulated splicing
A research team led by Dr. XUE Yuanchao from the Institute of Biophysics, Chinese Academy of Sciences, has developed a new method for global profiling of in situ RNA-RNA contacts associated with a specific RNA-binding protein (RBP) and revealed positional mechanisms by which PTBP1-associated RNA loops regulate cassette exon splicing.
This study was published online in Molecular Cell on Mar. 22.
In eukaryotes, a same pre-mRNA can produce multiple protein isoforms to execute similar or different biological functions through alternative splicing. Several long-standing models proposed that RBPs may regulate alternative splicing by modulating long-range RNA-RNA interactions (RRI). However, direct experimental evidence is still lacking.
To fill the knowledge gap, researchers created a capture RIC-seq (CRIC-seq) method to map specific RBP-associated RRIs by leveraging the principle of RIC-seq technology and immunoprecipitation-mediated enrichment. Notably, RIC-seq technology was developed by Dr. XUE's group in 2020 for global profiling of all the expressing RBP-mediated RRIs, but not a specific one.
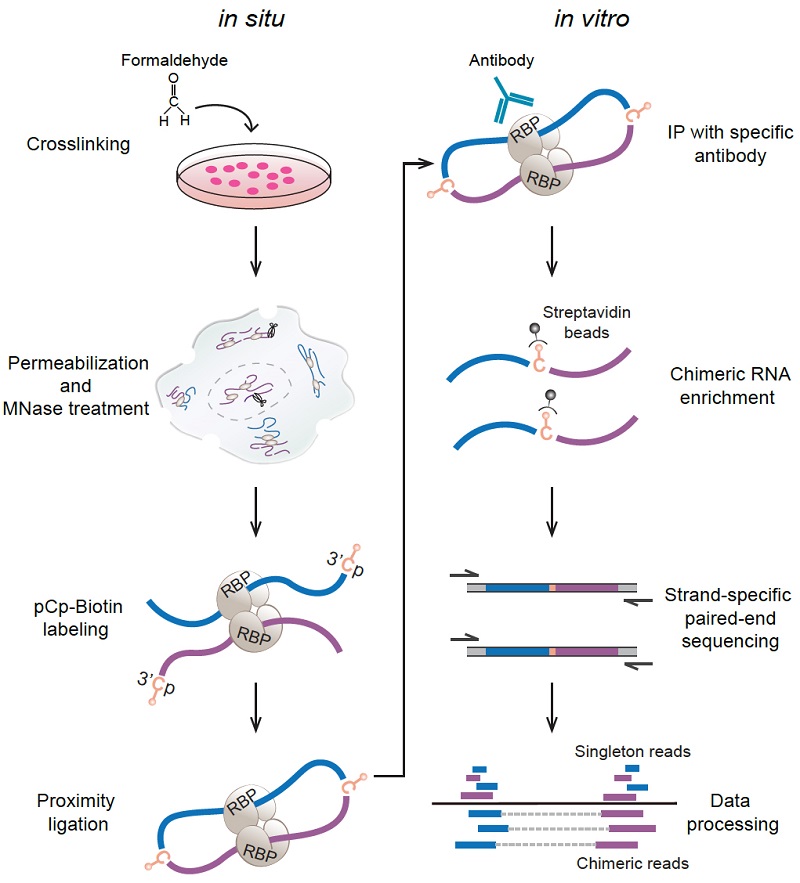
Figure 1. Diagram of the CRIC-seq method
Using the CRIC-seq method and the homemade data analysis pipeline, researchers obtained high-confidence RRIs mediated by PTBP1, hnRNPA1, or SRSF1 and generated functional 3D RNA maps to investigate their positional mechanisms from a new angle of RNA spatial conformation.
Similar to hnRNPA1, but unlike SRSF1, researchers found that PTBP1-mediated RNA loops tended to be enriched in one side of the intron when enhancing the splicing of cassette exons. In contrast, when repressing splicing, PTBP1-mediated RNA loops tended to span across the cassette exons. These findings demonstrate that PTBP1 can regulate cassette exon splicing via mediating specific RNA loops in a position-dependent manner.
According to minigene-based analysis, researchers found that only wild-type (WT) PTBP1, but not the C23S mutant PTBP1, can form a homodimer to rescue the corresponding splicing changes after PTBP1 knockdown. Furthermore, the CRISPR-dCasRx-mediated tethering assay confirmed that engineered RNA loops by WT PTBP1 can modulate the usage of non-PTBP1-regulated cassette exons. These results demonstrate that PTBP1 dimerization can drive RNA looping to modulate splicing.
In addition, by integrating cancer-related splicing quantitative trait loci (sQTLs) with PTBP1-associated RRIs, researchers identified 121 sQTLs that potentially affect splicing by altering RNA loops. Unexpectedly, one sQTL located in the intronic loops of HERPUD2 can induce exon 7 skipping, thus generating a short isoform to promote the proliferation of HeLa cells. These results demonstrated how cancer-related non-coding mutations modulate RNA spatial conformation to promote tumor cell growth.
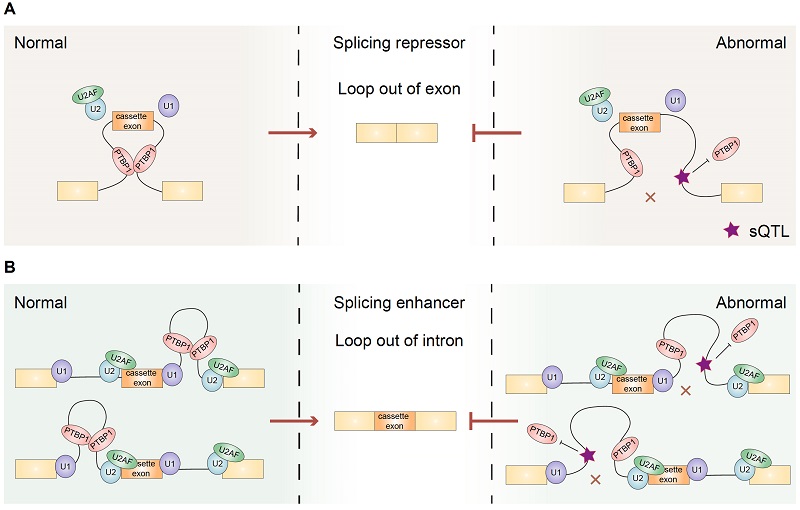
Figure 2. PTBP1-associated RNA loops in splicing regulation and tumor cell growth
This study provides a powerful method for the global mapping of specific RBP-associated RRIs and a conceptual framework for understanding gene regulation and disease occurrence from a new angle of RNA spatial conformation.
Article link: https://doi.org/10.1016/j.molcel.2023.03.001
Contact: XUE Yuanchao
Institute of Biophysics, Chinese Academy of Sciences
Beijing 100101, China
Email: ycxue@ibp.ac.cn
(Reported by Dr. XUE Yuanchao's group)

