Dr. SUN Fei's Group Publishes a Review Article on the Structure of the Nuclear Pore Complex in a Special Issue of the Journal of Molecular Biology
The nuclear pore complex (NPC) serves as the sole gateway for nucleocytoplasmic transport. Due to its complex composition and important biological functions, successfully analyzing the structure of NPC has long been considered the "Holy Grail" of structural biology. With recent advances in cryo-electron microscopy (Cryo-EM) and artificial intelligence (AI) technologies, near-complete skeletal structures of NPCs have been resolved through the collaborative efforts of many outstanding scientists. This achievement marks a significant milestone in the structural analysis of this massive protein machinery. On May 5, 2023, Dr. SUN Fei's research group from the Institute of Biophysics of the Chinese Academy of Sciences published a cover review article in the Journal of Molecular Biology's special issue, New Frontier of Cryo-Electron Microscopy Technology, titled "Cryo-electron Microscopy Reveals the Structure of the Nuclear Pore Complex." This review comprehensively introduces our latest understanding of the structure and assembly of the NPC, reviews the historical process of using cryo-EM technology to improve the resolution of NPC structural analysis and developements from in vitro to in vivo studies, and discusses the future direction of NPC structural studies.
The NPC is a massive complex with eightfold symmetry, assembled from approximately 30 different types of nucleoporins (Nups), and that it can be further divided into multiple subcomplexes. The stable subcomplexes include the Y complex and the inner ring subcomplex (IRC). The Y complex, the structural core of the outer ring, is mainly composed of the β-propeller and α-helical stack domains. The IRC contains the key component, the central channel nucleoporin heterotrimer (CNT), which is composed of Nsp1, Nup49, and Nup57, and other proteins including Nup155, Nup93, and Nup205/Nup188. Aside from a brief review of the structural studies covering these subcomplexes, the article further introduces the structural domain composition and various assembly methods of these subcomplexes. For example, although the NPC is a channel component, few subcomplexes have structural domains that interact with lipids, and most Nups do not make direct contact with lipids.
The article systematically reviews the NPC research history, with a focus on structural studies based on cryo-EM technology, including cryo-electron tomography (Cryo-ET) and cryo-electron single particle analysis (Cryo-SPA). The resolution obtained from NPC structural research has continuously improved alongside the technological advancements of cryo-EM methods, starting from early, low-resolution (>6 nm) studies to now obtaining near-atomic resolution of structures in some NPC regions (Figure 1). The emergence of highly accurate AI-based protein structure prediction tools, such as AlphaFold and RoseTTAFold, has greatly assisted in constructing NPC structural models more quickly and comprehensively than before, deepening our understanding of NPC assembly and its complete structure. Additionally, with an increasing number of articles focusing on the in situ functional state of NPC in the past decade, researchers have gradually realized the importance of functional state and in situ structural studies to understand the physiological and biochemical functions of the NPC. New and intriguing research questions have also emerged, such as how substances are transported between the cytoplasm and nucleus through the NPC, the role of the nuclear basket, and how it interacts with the transport cargo. We believe that these questions will soon be answered with the rapid development of cryo-EM and AI-related technologies, leading to a deeper understanding of the foundations of life.
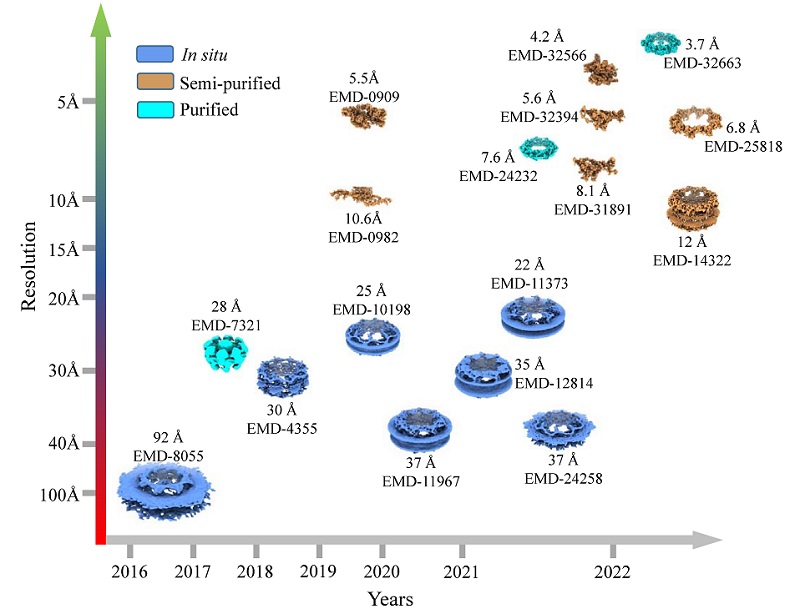
Figure 1. The resolution of the nuclear pore complex structure has continuously improved in recent years.
Researchers ZHU Yun and SUN Fei from Dr. SUN Fei's research group at the Institute of Biophysics of the Chinese Academy of Sciences are the corresponding authors of this review article, while Dr. TAI Linhua and Ph.D. candidate Guoliang Yin serve as co-first authors. In 2022, this research team published a paper titled "8 Å structure of the outer rings of the Xenopus laevis nuclear pore complex obtained by cryo-EM and AI" in Protein & Cell. The study obtained an 8 Å, near isotropic cryo-EM density map of the outer rings of the NPC. With assistance from AlphaFold2, the authors built the structures of all nucleoporin proteins and performed manual modeling to obtain a near complete structural model of the outer rings of NPC. The model revealed the spatial positions and interactions of multiple subunits, allowing for analysis of the structure of the NPC outer rings and laying the foundation for a comprehensive understanding of the overall structure and function of the NPC.
This review article is included in the Journal of Molecular Biology's special issue, New Frontier of Cryo-Electron Microscopy Technology. The editorial board of this special issue consists of Prof. SUN Fei and Prof. ZHANG Xinzheng from the Institute of Biophysics of the Chinese Academy of Sciences, and Assistant Prof. ZHANG Kai from Yale University. The special issue comprises four research articles and five review articles introducing the research process and latest advances in cryo-EM technology, emphasizing its importance in biomolecular structure studies and its broad applications in fields including drug development. Rapid progress is being made in the sample preparation, three-dimensional reconstruction algorithms, local resolution assessment, model building, and structural dynamics analysis of cryo-EM technology. Furthermore, the emergence of AI technology has significantly advanced the modeling of medium-to-low-resolution density maps, opening up new possibilities for developing cryo-EM technology (Figure 2).

Figure 2. Cover of the special issue, highlighting the crucial role of integrating artificial intelligence and cryo-electron microscopy technologies for the future structural analysis of macromolecular protein complexes.
Link to the original article:
https://www.sciencedirect.com/science/article/pii/S0022283623001079
Link to the special issue:
https://www.sciencedirect.com/journal/journal-of-molecular-biology/vol/435/issue/9
Contact: ZHU Yun
Institute of Biophysics, Chinese Academy of Sciences
Beijing 100101, China
Email: zhuyun@ibp.ac.cn
(Reported by Dr. SUN Fei's group)

