Comments on a proposed method for direct visualization of in-situ structural features and conformations in cryo-electron tomograms
On May 22, 2023, Nature Communications published a research paper by researchers lead by Prof. ZHU Ping at the Institute of Biophysics, Chinese Academy of Sciences. The paper by Zhang et al., entitled "A method for restoring signals and revealing individual macromolecule states in cryo-ET, REST" proposed a deep learning strategy based method for the REstoring the Signal in Tomograms, which was named REST. REST method aims to restore the missing information in cryo-electron tomography (cryo-ET) and enables direct observation and identification of the in-situ structural features and dynamic changes of target proteins with high signal-to-noise ratio from cryo-ET three-dimensional (3D) reconstruction results.
Cryo-ET can obtain in-situ 3D structures of biomolecules in cellular and tissue samples at nanoscale resolution. This information provides valuable insights and complementary information to structural and cell biology in terms of research scale and structural resolution. However, there are still several limitations to this technique. First, given the vulnerability of the biological frozen samples to electron radiation damage, low-dose imaging is required during the data acquisition process, where multiple images are taken from different angles of the same target region. This low-dose imaging results in electron tomograms with extremely low signal-to-noise ratio, posing significant challenges in subsequent image processing, 3D structure reconstruction, and identification of target biomolecules. Second, due to constraints such as the sample stage and imaging geometry during the data acquisition process, images can only be collected within a tilt angle range ± 60-70°. The absence of high tilt angle information leads to incomplete information in Fourier space, resulting in a phenomenon known as the "missing wedge" in electron tomograms. This causes significant resolution anisotropy in the 3D reconstruction of electron tomograms (with lower resolution along the Z-axis compared to the X-Y axes) and the formation of severe artifacts. These two factors greatly hinder the identification and analysis of the target molecules of interest and their features in cryo-ET 3D reconstructions.
In recent years, deep learning techniques have made significant advancements in areas such as denoising, and target and feature identification. However, the extremely low signal-to-noise ratio and irreversible information loss in cryo-ET make it difficult for researchers to obtain the real information of target particles, which can be used as ground truth and is essential for the deep learning process. Consequently, the application of neural networks and deep learning techniques for the identification of target macromolecules in electron tomograms pose significant challenges.
Considering the issue of the heavy reliance of the training process of deep learning on the training dataset, and difficulty in obtaining the "ground truth" for training neural networks in cryo-ET, Zhang et al. proposed and implemented two training strategies in their recent research paper (Figure 1). Strategy 1 selected a small subset of particles from the original data and performed sub-tomogram averaging on these particles. The results were then used as the "ground truth" and paired with the original particles for training. This approach established a mapping relationship between the original data and the averaged results, allowing the knowledge transfer of information obtained from a limited number of original particles to recover more information from the original particles. In Strategy 2, the researchers simulated the low signal-to-noise ratio and macromolecular structural heterogeneity in real data by adding different levels of noise and dynamic conformational changes to the high-quality "ground truth" density maps (e.g., 3D density maps generated using atomic structures of the target protein). The simulated low-quality density maps with high noise and dynamic changes were then paired with the high-quality density maps (ground truth) to form the training dataset. Once these training datasets and deep learning strategies were established, the researchers utilized deep learning networks to learn from and train on the datasets. The trained models and acquired knowledge were then applied to the original data to recover the information of target protein particles.
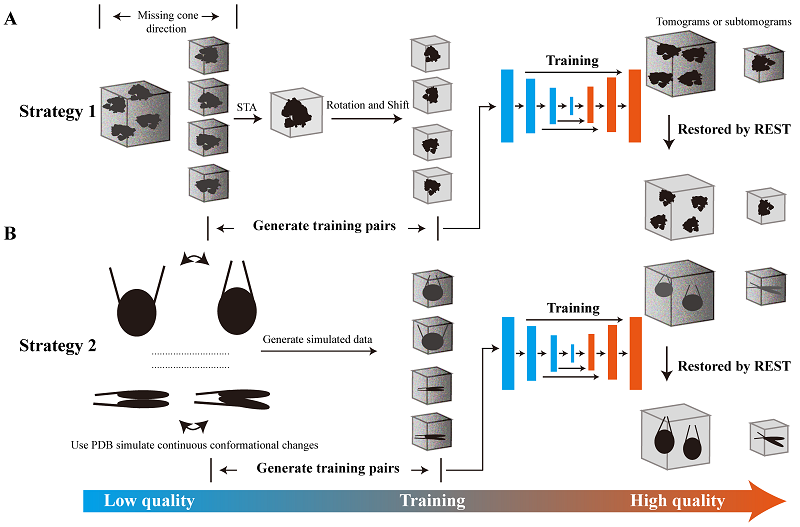
Figure 1. Workflow and training strategies of REST
By utilizing the REST method with these strategies, Zhang et al. were able to improve the signal-to-noise ratio by one-to-two orders of magnitude in the cryo-ET 3D reconstructions of target proteins. This improvement greatly enhanced the efficiency and accuracy of identifying target protein molecules and their features. Interestingly, REST Strategy 2 obtained multiple new conformational states of the target protein that were not included in the training dataset. This greatly enhanced the ability to identify the dynamic structures of target molecules in cellular in-situ structures, which is crucial for identifying and visualizing the in-situ structural variations of target biomolecules with dynamic conformational changes.
In summary, REST is a novel approach to directly visualize and interpret the 3D structures and conformational changes of target proteins in cryo-ET reconstructions with high signal-to-noise ratio. This method has broad application prospects and value in tasks related to cryo-ET such as recovering clear signals of target proteins (e.g., identifying and picking particles from noisy backgrounds), segmenting target features, identifying the dynamic or flexible structures of target proteins, obtaining density without missing information as an initial model, and assisting sub-tomogram averaging.
ZHANG Haonan, a Ph.D. student, and LI Yan, an associate researcher of the ZHU Ping's group at the Institute of Biophysics, Chinese Academy of Sciences are the co-first authors of the Nature Communications paper. ZHU Ping is the corresponding author of the paper. This research work was supported by the National Natural Science Foundation of China, the Key Research and Development Program of Chinese Ministry of Science and Technology, and the Strategic Pioneering Science and Technology Special Project of the Chinese Academy of Sciences (Class B).
Original article link:
https://www.nature.com/articles/s41467-023-38539-w
Contact: ZHU Ping
Institute of Biophysics, Chinese Academy of Sciences
Beijing 100101, China
Email: zhup@ibp.ac.cn
(Reported by Dr. ZHU Ping's group)

