Scientists Reveal Mechanism Behind Preferential Recognition of H2A.Z Nucleosome During DNA Replication Initiation
In higher eukaryotes, DNA replication ensures the precise transmission of genetic information from parental DNA to progeny, and the correct selection of replication origins is a critical step in this process. A very recent study has shown that nucleosomes containing histone variant H2A.Z recruits SUV420H1, a histone H4 lysine 20 specific methyltransferase. Moreover, SUV420H1 promotes the H4K20 dimethylation which can recruit the origin recognition complex to fulfill the selection and licencing of the replication origins.
On August 2, 2023, the research group of Prof. ZHOU Zheng from the Institute of Biophysics, Chinese Academy of Sciences, in collaboration with Prof.LI Guohong and Prof. ZHU Ping from the same institute, and Prof. LONG Haizhen from the Shenzhen Bay Laboratory, published a research paper titled "Structural Insight into H4K20 Methylation on H2A.Z-Nucleosome by SUV420H1" in Molecular Cell. The study reported the high-resolution cryo-electron microscopy structure of the human SUV420H1 bound to H2A.Z nucleosome, revealing the molecular mechanism of SUV420H1's preferential recognition of H2A.Z nucleosome and catalysis of H4K20me2 (Figure 1).
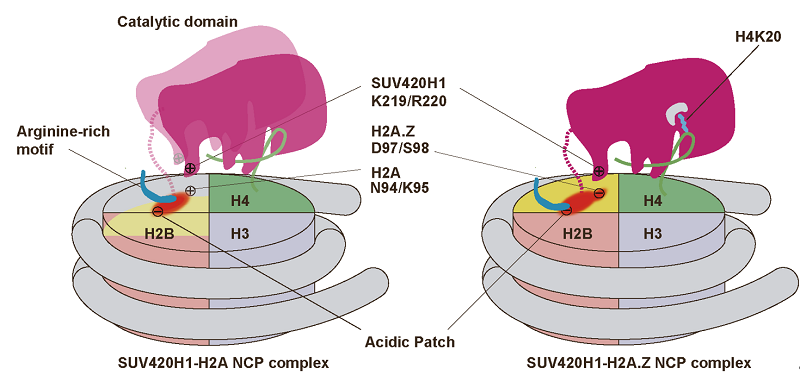
Figure 1: Schematic view of SUV420H1 structures bound to H2A.Z nucleosome
The researchers replaced H4 to lysine 20 residue with a S-ethyl-L-cysteine (Ecx), a modification which stabilizes the SUV420H1-H2A.Z nucleosome interaction, and led to determination of the complex structure at a 3.2 Å resolution. The structure revealed SUV420H1's interactions with the H4 N-terminal tail, the DNA, and the acidic patch of nucleosome. Particularly, the H4 N-terminal tail adopts a lasso-like structure that precisely positions H4K20 into the catalytic center of SUV420H1 (Figure 1B). Notably, the researchers identified an SUV420H1 KR-loop in close proximity to the H2A.Z-specific residues D97/S98 (corresponding to H2A residues N94/K95). The KR-loop plays a role in conferring SUV420H1's preference for the H2A.Z nucleosome in vitro. And its mutations decrease the cellular level of H4K20me2, the DNA replication initiation, and the growth rate of cell. These findings are in full agreement with earlier studies, which indicated that H2A.Z D97/S98 governed SUV420H1's preference for the H2A.Z nucleosome.
H2A.Z is a key histone variant involved in numerous biological processes. Prof. ZHOU Zheng's research group is dedicated to investigating the mechanisms underlying H2A.Z recognition and function. This study revealed how SUV420H1 preferentially recognizes the H2A.Z nucleosome to deposite the H4K20me2 marks, and provided novel insights into regulation of DNA replication initiation. Most importantly, this study, for the first time, unravels the mechanism of preferential recognition of H2A.Z in the context of the nucleosome and broaden our understanding of histone variants.

Cover: Huang et al. reveal the intricate mechanism by which the methyltransferase SUV420H1 preferencially recognizes H2A.Z-containing nucleosomes to catalyze H4K20me2 modification. The artwork showcases SUV420H1, portrayed as a whimsical elf, skillfully illuminating the histone marker, represented by a magical wand, amidst the chromatin landscape. This study dissect the role of H2A.Z and SUV420H1 in replication origin selection and licensing that ensure the accurate initiation of DNA replication.
Prof. ZHOU Zheng, Prof. LI Guohong, Prof. ZHU Ping, and Prof. LONG Haizhen from the Institute of Biophysics, Chinese Academy of Sciences, are co-corresponding authors of this research. Dr. HUANG Li, a postdoctoral researcher, and Dr. WANG Youwang, and Prof. LONG Haizhen are co-first authors of this paper. The research was supported by grant from the National Natural Science Foundation of China, the Department of Science and techanology, and the Chinese Academy of Sciences, and support from the Protein Science Platform and Biological Imaging Center of the Institute of Biophysics.
Article link: https://doi.org/10.1016/j.molcel.2023.07.001
Contact: ZHOU zheng
Institute of Biophysics, Chinese Academy of Sciences
Beijing 100101, China
Email: zhouzh@ibp.ac.cn
(Report by Prof. ZHOU zheng's group)

