YE Keqiong Laboratory identified new rules governing substrate recognition by archaeal C/D guide RNAs
On Nov 30, 2023, the YE Keqiong Laboratory at the Institute of Biophysics, Chinese Academy of Sciences (CAS), published a paper entitled "Complicated target recognition by archaeal box C/D guide RNAs" in Science China Life Sciences. This study revealed the landscape of 2'-O-methylation modification in the archaeon Sulfolobus islandicus and identified new targeting rules employed by C/D RNA.
2'-O-methylation is one of the most frequent modifications in RNA and modulates the structure, function and stability of RNA. C/D RNAs are universally present in archaea and eukaryotes and guide the site-specific 2'-O-methylation of substrate RNAs. Classic C/D RNAs recognize substrates through base pairing interaction and selects a site located 5 nt upstream the box D/D' motif for modification, known as the D+5 rule. Although archaeal C/D RNAs have been extensively studied by in vitro biochemical and structural approaches, their targets and substate recognition mechanisms have not been systemically investigated in vivo.
The study systemically mapped 2'-O-methylation in rRNAs, tRNAs, and abundant small RNAs and profiled C/D RNAs in the crenarchaeon Sulfolobus islandicus. The targets of C/D RNAs were assigned by analysis of base-pairing interactions, in vitro modification assays, and gene deletion experiments, revealing a complicated landscape of C/D RNA-target interactions. The study found that C/D RNAs widely use dual antisense elements to target adjacent sites in rRNAs, accounting for 79% of 2'-O-methylation. One of substrates in dual-targeting is commonly weakly bound. Biochemical assays showed that such dual binding can enhance the modification of the weakly bound site. The study found two C/D RNAs with double-specificity that can guide modification of two consecutive sites with the same antisense element. Double-specificity guides were also found in eukaryotic C/D RNAs, but exclusively present at the D' guide. On contrast, they can be present in the D or D' guide in archaea. Finally, the study revealed for the first time that C/D RNAs can guide modification at a single non-canonical site, including position 4, 6 and 7 upstream of box D/D'. Overall, this study reveals the landscape of 2'-O-methylation in an archaeon and unexpected targeting rules employed by C/D RNA.
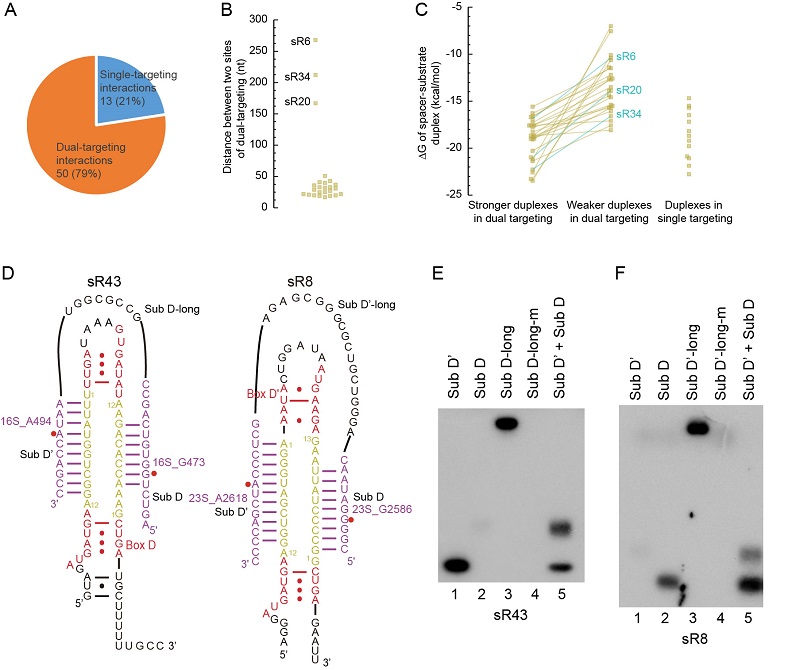
Figure. C/D RNAs targeting dual adjacent sites of rRNAs
This work was conducted at the Institute of Biophysics, CAS. Drs. WANG Jiayin and WU Songlin are the co-first authors and Prof. YE Keqiong is the corresponding author of the paper. This work was supported by National Natural Science Foundation of China, Strategic Priority Research Program of Chinese Academy of Sciences and National Key R&D Program of China.
Paper hyperlink: https://doi.org/10.1007/s11427-022-2412-3
Contact: Ye Keqiong
Institute of Biophysics, Chinese Academy of Sciences
Beijing 100101, China
Email: yekeqiong@ibp.ac.cn
(Reported by Prof. YE Keqiong’s group)

