Keqiong Ye Laboratory reveals landscape of RNA pseudouridylation in an archaeon
On Feb 20, 2024, the YE Keqiong Laboratory at the Institute of Biophysics, Chinese Academy of Sciences (CAS), published online a paper entitled "Landscape of RNA pseudouridylation in archaeon Sulfolobus islandicus" in Nucleic Acids Research. This study revealed a first global view of pseudouridine distribution and synthesis in an archaeon.
Pseudouridine is one of the most frequent modifications present in rRNA, tRNA, snRNA and mRNA and modulates their structure and function. Pseudouridine is enzymatically synthesized through post-transcriptional conversion of uridine by pseudouridine synthases (PUSs), which can be divided into six families. Cbf5, belonging to the TruB family, is a special PUS that associates with other three proteins and a distinct H/ACA RNA to form an RNA-protein complex (RNP) and uses the H/ACA RNA to recognize substrates. The other PUSs are all stand-alone proteins that directly recognize substrates. The distribution and synthesis of pseudouridine have been well studied in several eukaryotic and bacterial model organisms, but not in the third life kingdom archaea.
The study systemically mapped pseudouridine in rRNAs, tRNAs and abundant small RNAs and profiled Cbf5-associated H/ACA RNAs in the model archaeon Sulfolobus islandicus. These modifications were assigned to the responsible enzymes through genetic deletion and in vitro modification assays. The study found that the pseudouridylation machinery in this organism consists of the stand-alone enzymes aPus7 and aPus10, and six H/ACA RNA-guided enzymes that account for all identified pseudouridines. These H/ACA RNAs guide the modification of all eleven sites in rRNAs, two sites in tRNAs and two sites in CRISPR RNAs. One H/ACA RNA shows exceptional versatility by targeting eight different sites. aPus7 and aPus10 are responsible for modifying positions 13, 54 and 55 in tRNAs. In addition, the study identified four atypical H/ACA RNAs that lack the lower stem and the ACA motif and confirmed their function both in vivo and in vitro. Intriguingly, atypical H/ACA RNAs can be modified by Cbf5 in a guide-independent manner.
In sum, the research provides the first global view of pseudouridylation in archaea and reveals unexpected structures, substrates and activities of archaeal H/ACA RNPs.
This work was conducted at the Institute of Biophysics, CAS. LI Yuqian is the first author, Prof. YE Keqiong is the corresponding author of the paper and the other authors include Dr. WU Songlin. This work was supported by National Natural Science Foundation of China, Strategic Priority Research Program of Chinese Academy of Sciences and National Key R&D Program of China.
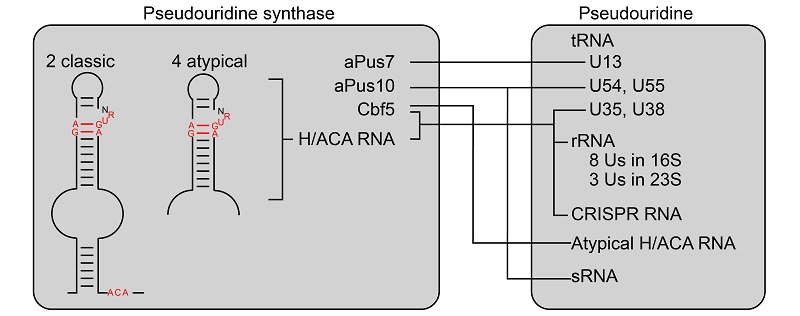
Figure. Distribution and synthesis of pseudouridine in Sulfolobus islandicus
Article link: https://doi.org/10.1093/nar/gkae096
Contact: YE Keqiong
Institute of Biophysics, Chinese Academy of Sciences
Beijing 100101, China
Email: yekeqiong@ibp.ac.cn
(Reported by Prof. YE Keqiong's group)

