Scientists Develop New Method for Cryo-electron Tomography Particle Picking
Scientists from the Center for Biological Imaging, Institute of Biophysics, Chinese Academy of Sciences, collaborated with scientists from State Key Laboratory of Multimodal Artificial Intelligence Systems, Institute of Automation, Chinese Academy of Sciences, to empower in-situ structural biology with artificial intelligence technology, proposing a fast and accurate particle picking method based on weakly supervised deep learning, named DeepETPicker. The study was published in Nature Communications.
Cryo-electron tomography (cryo-ET) provides a powerful tool for visualizing macromolecular complexes under native conformations at sub-nanometre resolutions and for revealing their spatial and organizational relationships. This provides new mechanistic insights into key cellular processes and new possibilities for applications such as drug discovery. As biological samples are very sensitive to radiation damage, the native resolution of cryo-ET is limited to ~2-5 nm given the dose of imaging electrons that can be tolerated. This resolution is insufficient for studying the structures and functions of macromolecular complexes. Subtomogram averaging (STA) is commonly used to obtain higher-resolution structures by aligning and averaging large numbers of particles of the same macromolecular complexes. Particle picking in this technological process, i.e., locating recognition, is a crucial step. However, the application of existing automatic picking methods is limited by various factors such as the large amount of manual annotation, high computational costs, and poor particle quality.
DeepETPicker, as introduced, requires only a small amount of manual annotation of particles for training to achieve fast and accurate three-dimensional particle automatic picking. DeepETPicker optimally simplifies labels to replace real labels and adopts a more efficient model architecture, richer data augmentation techniques, and overlap partitioning strategies to enhance the model's performance with small training sets. To improve particle localization speed, DeepETPicker employs GPU-accelerated average pooling and non-maximum suppression post-processing operations, achieving a several-fold increase in picking speed compared to existing clustering post-processing methods.
In a comprehensive evaluation of the performance of particle picking quality based on six quantitative metrics, DeepETPicker can achieve fast and accurate particle picking on both simulated and real datasets. Its overall performance is significantly better than existing methods, and the resolution of the structural reconstruction of biomacromolecules reaches the level achieved by manual expert particle picking for structure reconstruction, further demonstrating the practical value of DeepETPicker in in-situ high-resolution structural analysis.
Currently, to facilitate user usage, the research team has launched open-source software with simple operation and user-friendly interface to assist users in completing image preprocessing, particle annotation, model training, and inference operations.
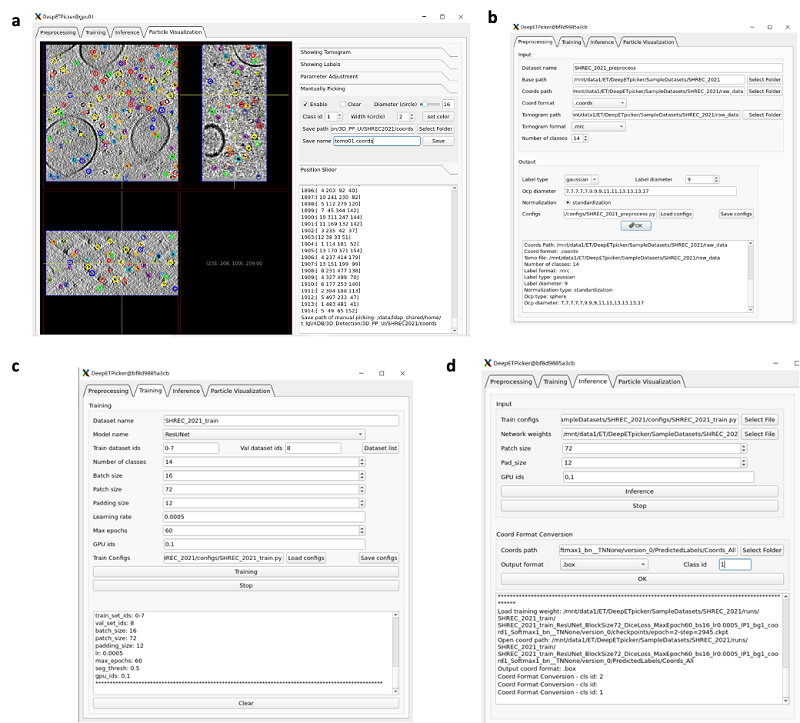
Fig 1. The User Graphical Interface of DeepETPicker
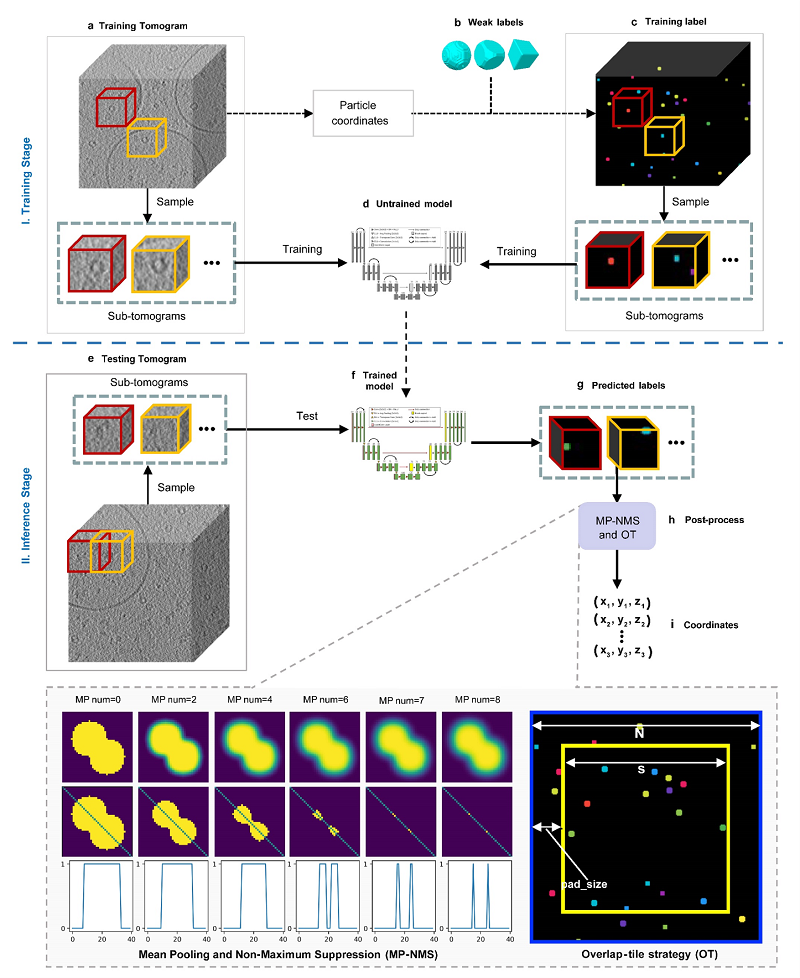
Fig 2. The workflow for picking particles from cryo-electron tomograms using DeepETPicker
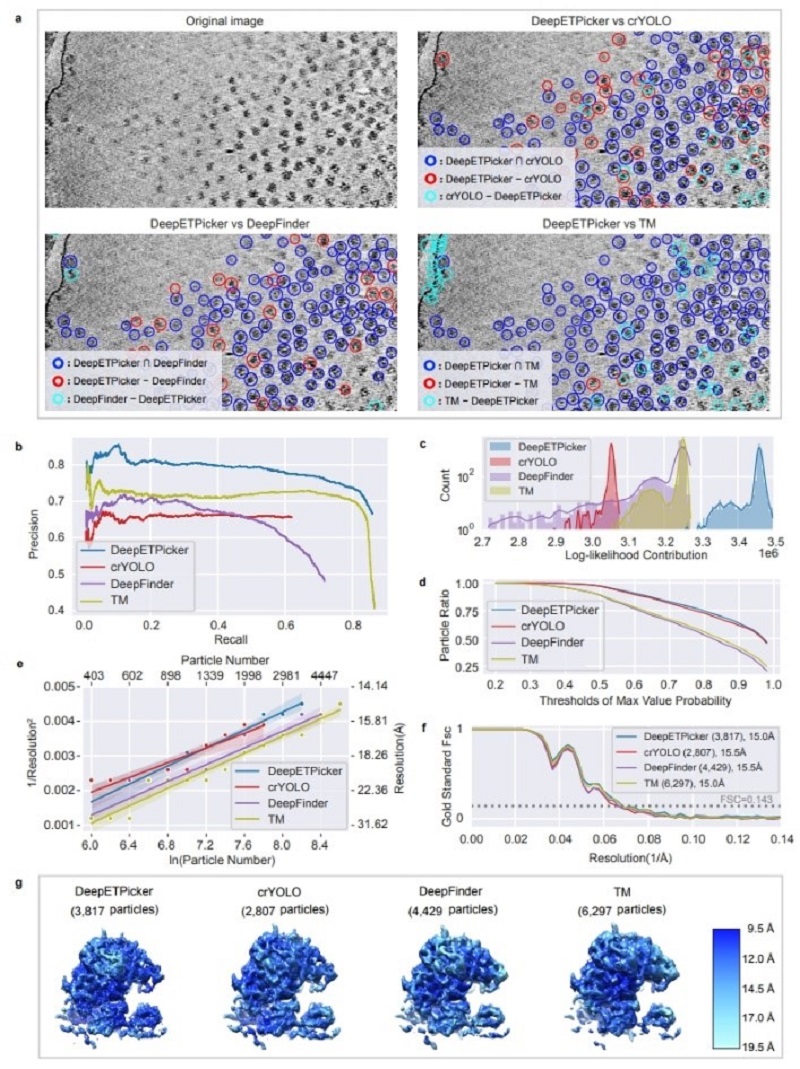
Fig 3. evaluates the particle picking performance of DeepETPicker on the EMPIAR-10045 experimental dataset using quantitative metrics
Article link: https://www.nature.com/articles/s41467-024-46041-0
Code availability: https://github.com/cbmi-group/DeepETPicker
Contact: NIU Tongxin
Institute of Biophysics, Chinese Academy of Sciences
Beijing 100101, China
Email: niutx@ibp.ac.cn
(Reported by Center for Biological Imaging)

