YE Keqiong Laboratory develops a high-resolution technique to dissect ribosomal RNA processing mechanism
On July 12, 2024, the YE Keqiong Laboratory at the Institute of Biophysics, Chinese Academy of Sciences (CAS), published online a paper entitled "High-resolution landscape of ribosomal RNA processing and surveillance" in Nucleic Acids Research. This study developed a high-resolution technique to profile rRNA processing intermediates and discovered several new mechanisms in rRNA processing and surveillance.
Ribosomal RNAs (rRNAs) are the most abundant RNA and make up approximately 80% of total RNA in eukaryotic cells. Synthesis of rRNA is a fundamental, complicated and conserved cellular activity where precursor rRNAs are processed by a series of endo- and exo-ribonucleases to generate mature 18S, 5.8S and 25S rRNA. In addition, aberrant processing intermediates are degraded by the nuclear RNA surveillance system. The major steps of rRNA processing and surveillance have long been established in yeast, but there are still steps that are not or incompletely characterized.
Traditional methods to detect rRNA processing intermediates, such as Northern blot, are limited in resolution and sensitivity, and particularly difficult for analyzing low-abundance intermediates with continuous ends. In this study, the authors developed a high-throughput method comprising of RNA circularization, targeted amplification, and deep sequencing (CircTA-seq) to profile rRNA processing intermediates at single-molecule and single-nucleotide resolution.
By analyzing the complex population of pre-rRNA and its disturbance by 15 mutants with unprecedented precision, sensitivity and resolution, the study obtained major mechanistic understanding into rRNA processing and surveillance. The 5.8S rRNA has a long and short form of the 5' end which are currently considered to be generated through two different pathways. The authors found that the long form is slowly converted into the short form during RNA processing and proposed a unified model to account for their biogenesis. In addition, the study found that the initial 3' end processing of 5.8S rRNA involves trimming by Rex1 and Rex2 and Trf4-mediated polyadenylation, which subsequently activates the exosome. The study also shows that the 3' end of 25S rRNA is formed by sequential digestion by four Rex proteins.
Pre-rRNA processing is monitored by the nuclear RNA surveillance system. Aberrant processing intermediates are appended with a short polyadenosine (poly(A)) tail that activates the nuclear exosome for subsequent degradation. The study determined precise polyadenylation profiles for pre-rRNAs and show that the degradation efficiency of 20S pre-rRNA critically depends on poly(A) lengths and degradation intermediates released from the exosome are often extensively re-polyadenylated. The study also identified novel intermediates from 5' degradation of aberrant 18S rRNA precursors.
In sum, this study invented a high-resolution technique to profile rRNA processing intermediates and identified several novel mechanisms in rRNA processing and surveillance, greatly improving the classic rRNA processing pathway.
This work was conducted at the Institute of Biophysics, CAS. Dr AN Weidong is the first author, Prof. YE Keqiong is the corresponding author of the paper and the other authors include YAN Yunxiao. This work was supported by National Natural Science Foundation of China, Strategic Priority Research Program of Chinese Academy of Sciences and National Key R&D Program of China.
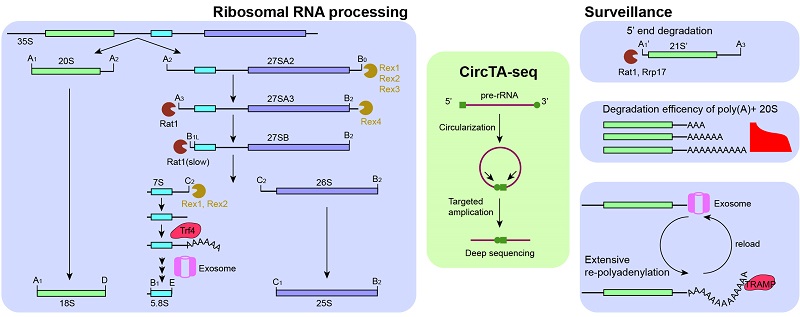
Figure. CircTA-seq analysis uncovered new mechanisms in yeast rRNA processing and surveillance
(Image by YE Keqiong's group)
Article link: https://doi.org/10.1093/nar/gkae606
Contact: YE Keqiong
Institute of Biophysics, Chinese Academy of Sciences
Beijing 100101, China
E-mail: yekeqiong@ibp.ac.cn
(Reported by Prof. YE Keqiong's group)

