Pramel15 Facilitates Zygotic Nuclear DNMT1 Degradation and DNA Demethylation
In mammals, the zygote undergoes DNA methylation reprogramming, resetting the methylation status inherited from parental genome to prepare for subsequent tissue differentiation and embryonic development. Aberrate DNA methylation reprogramming can lead to issues such as embryonic development defects and physiological abnormalities in the offspring. During early embryonic development, the genome undergoes global DNA demethylation, primarily driven by replication-dependent passive demethylation. The cytoplasmic retention of the key DNA methylation maintenance enzyme DNMT1 (DNA methyltransferase 1) and its cofactor UHRF1 in oocytes and early embryos is considered a major reason for passive DNA demethylation during this period. The TET (Ten-eleven translocation) family of dioxygenases can sequentially oxidize 5-methylcytosine (5mC) to produce 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC), and 5-carboxylcytosine (5caC). These oxidized products can either be gradually loss during DNA replication or restored to cytosine through base excision repair, the latter is known as replication-independent active DNA demethylation. TET3, the only TET protein expressed in oocytes, plays a role in specific region-targeted active demethylation that also aids in the DNA methylation reprogramming of early embryos. Deficiency of TET3 can lead to metabolic abnormalities in offspring, such as increased susceptibility to diabetes.
Although genome-wide DNA demethylation is a characteristic of DNA methylation reprogramming during early embryonic development, advanced studies have revealed that DNA methylation maintenance still occurs in some specific regions, such as imprinted genes and retroelements, during this stage. The maternal loss of DNMT1 results in the failure to maintain DNA methylation in these regions during reprogramming, leading to embryonic developmental abnormalities and lethality. Additionally, some studies have found that de novo DNA methylation occurs post-fertilization, dependent on DNMT1 and DNMT3A. DNA methylation reprogramming in early embryos involves several different regulations, and the roles of proteins directly involved, such as DNMT1 and UHRF1, have been well-studied. However, the mechanisms by which these proteins are regulated to function appropriately during DNA methylation reprogramming remain to be elucidated.
A research team led by Prof. ZHU Bing from the Institute of Biophysics, Chinese Academy of Sciences reported that the maternal factor Pramel15 can mediate DNMT1 degradation in the zygotic nucleus, fine-tuning nuclear DNMT1 protein abundance. This work was published in Nature Communications on August 25, 2024.
The team identified a novel DNA methylation regulator, Pramel15, from a mouse oocyte cDNA library. Overexpression of Pramel15 in somatic cells leads to global DNA demethylation by degrading DNMT1. Further investigation reveals that Pramel15 is a substrate recognition subunit of the Cullin5 E3 ubiquitin ligase complex, and mediates DNMT1 degradation via the ubiquitin-proteasome pathway. Notably, in Cullin5-deficient cells, Pramel15 can still degrade DNMT1 through other Cullin complexes.
To further understand the role of Pramel15 in early embryonic development, the researchers generated Pramel15 knockout mice. According to immunofluorescence staining and quantitative analysis of DNMT1 in Pramel15-deficient oocytes and early embryos, They found DNMT1 levels increased in the zygotic nucleus. The zygotes treated with the proteasome inhibitor MG132 and the Cullin ubiquitin ligase complex inhibitor MLN4924 also showed similar DNMT1 increases, while Pramel15-deficient zygotes did not exhibit further increases in nuclear DNMT1 levels after the drug treatment. This suggests that a Pramel15-mediated proteasome degradation pathway predominantly regulates DNMT1 in the zygote. At last, they performed whole-genome DNA methylation sequencing (WGBS) of oocytes, zygotes, and two-cell embryos. The results revealed that Pramel15 deficiency embryos have a stochastic gain of DNA methylation. In conclusion, Pramel15 regulates nuclear DNMT1 levels during DNA replication in zygotes, thus aiding DNA methylation reprogramming in early embryos.
Recently, several research groups have reported the mechanisms of cytoplasmic retention of DNMT1 and UHRF1 in oocytes and early embryos. They discovered that SCMC-related proteins NLRP14 and PADI6 regulate the cytoplasmic localization of UHRF1 and DNMT1, respectively. This study reveals that while DNMT1 and UHRF1 are transported to and restricted in the cytoplasm during early embryonic development, there is also a proteasome-mediated mechanism in the nucleus that controls DNMT1 protein levels, regulating the efficiency of DNA methylation reprogramming. This discovery highlights the complexity of DNA methylation reprogramming regulatory mechanisms in early embryo.
This work was funded by grants from the China Natural Science Foundation, the National Key R&D Program of China, the Chinese Academy of Sciences, the New Cornerstone Science Laboratory, and the Youth Innovation Promotion Association of the Chinese Academy of Sciences.
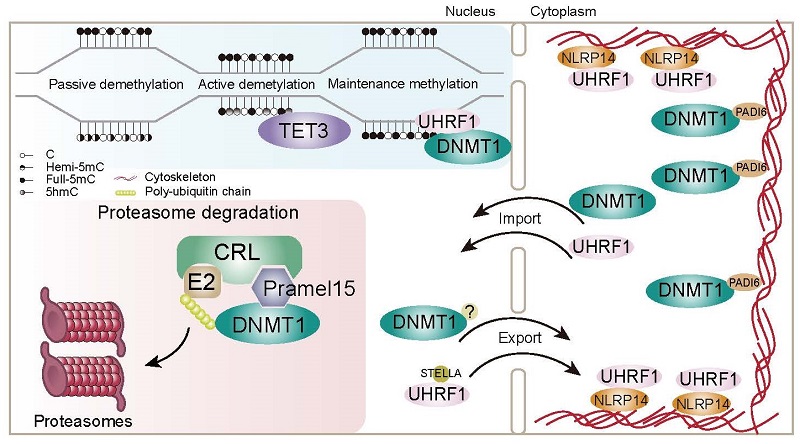
Fig. Regulatory mechanism of DNA methylation reprogramming during early embryonic development
(Image by ZHU Bing's group)
Article link:https://www.nature.com/articles/s41467-024-51614-0
Contact: ZHU Bing
Institute of Biophysics, Chinese Academy of Sciences
Tel: 010-64888832
E-mail: zhubing@ibp.ac.cn
(Reported by Prof. ZHU Bing's group)

