Scientists Review Molecular Ruler Techniques and Their Applications
The inherent flexibility and conformational dynamics of RNA molecules pose significant challenges for traditional structural techniques, such as X-ray crystallography, nuclear magnetic resonance (NMR), and cryo-electron microscopy, in resolving RNA structures and capturing RNA's dynamic changes.
Combining multiple methods and integrating experimental data with computational modeling can expand the research approaches and perspectives within structural biology.
On October 22, 2024, Prof. FANG Xianyang from the Institute of Biophysics, Chinese Academy of Sciences, reviewed three complementary molecular ruler techniques in Current Opinion in Structural Biology, including electron paramagnetic resonance (EPR), X-ray scattering interference (XSI), and fluorescence resonance energy transfer (FRET).
The article elaborates on the fundamental principles of these three techniques and demonstrates how to use the NaM-TPT3 unnatural base pair system to achieve site-specific labeling of long-chain RNA, allowing for nanoscale measurements of intra- and intermolecular distance distributions in RNAs.
This labeling method overcomes the limitations of traditional RNA labeling techniques concerning RNA length and labeling sites. It offers advantages such as high reaction efficiency, simplified purification, and the elimination of denaturing conditions during purification, thereby enabling the study of long-chain RNA structures and conformational dynamics using molecular ruler techniques.
By showcasing the application of various molecular ruler techniques in studying the structure and conformational dynamics of flaviviral RNAs, this research not only provides a more accurate understanding of the conformational changes in flaviviral RNAs but also identifies new targets and strategies for treating viral diseases.
Additionally, the application of these techniques holds promise for advancing RNA nanotechnology, offering more possibilities for future biomedical research.
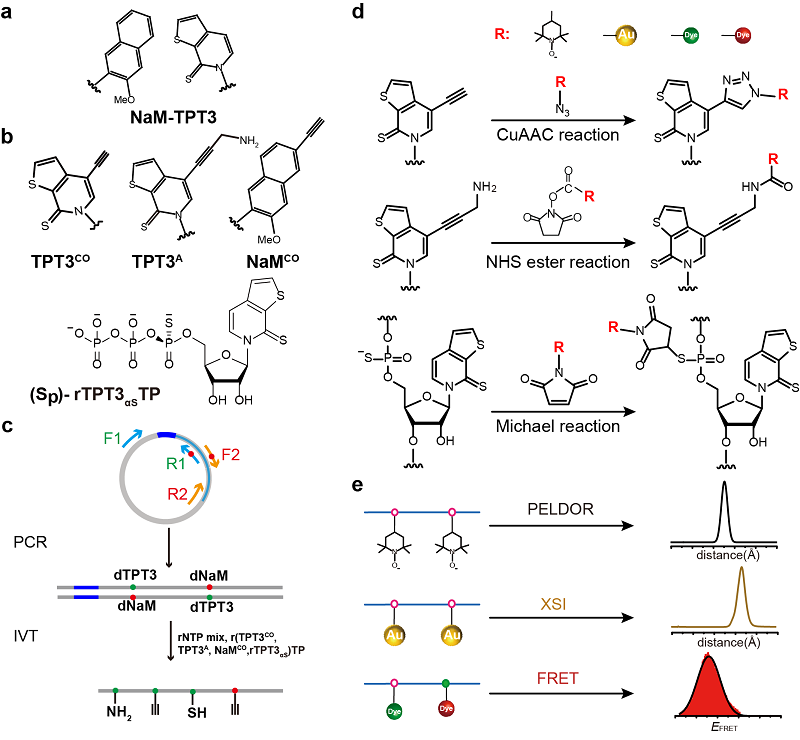
Fig. The NaM-TPT3 UBP-based site-specific labeling method for long-chain RNAs
(Image by FANG Xianyang's group)
Article link: https://doi.org/10.1016/j.sbi.2024.102944
Contact: FANG Xianyang
Institute of Biophysics, Chinese Academy of Sciences
Beijing 100101, China
Email: fangxy@ibp.ac.cn
(Reported by Prof. FANG Xianyang's group)

