Scientists uncovers a New Mechanism Regulating mRNA Poly(A) Tail in Early Embryos
Due to the absence of appreciable transcription during mammalian early embryonic development, the mRNA templates required for protein synthesis are primarily supplied by maternal mRNAs stored in oocytes. During the oocyte-to-embryo transition (OET), mRNA poly(A) tail length is tightly coupled with translational efficiency, poly(A) tail regulation is a crucial mechanism for selective translation in early embryos. However, the mechanisms determining and maintaining the length of maternal mRNA poly(A) tails have remained elusive.
On January 2, 2025, Dr. ZHU Bing's research group from the Institute of Biophysics, Chinese Academy of Sciences (CAS), in collaboration with Lu Falong's group from the Institute of Genetics and Developmental Biology, CAS, published their study titled "MARTRE family proteins negatively regulate CCR4-NOT activity to protect poly(A) tail length and promote translation of maternal mRNA" in Nature Communications. This study unveils a protective mechanism for the poly(A) tail of maternal mRNA in early embryos.
The team's earlier work compared mRNA degradation rates between mouse 2-cell-like cells (2CLCs) and embryonic stem cells (ESCs), finding increased mRNA stability in 2CLCs. This indicated a unique regulatory mechanism of mRNA degradation in 2CLCs. By identifying proteins associated with the poly(A) deadenylase CCR4-NOT complex, they discovered a poly(A) tail regulator selectively expressed in 2CLCs, which they named MARTRE1. Cellular and in vitro biochemical assays revealed that MARTRE1 inhibits CCR4-NOT complex activity. Expressing MARTRE1 in cells slowed the shortening of poly(A) tails and enhanced mRNA stability.
Homology analysis showed that Martre1 belongs to a previously underexplored gene family, which the researchers named Martre. Members of the Martre gene family are expressed exclusively during the zygotic genome activation in mouse oocytes and early embryos. Using a Martre gene family knockout mouse model, the team observed developmental delays in early embryos. Combining transcriptomics, translatomics, and third-generation sequencing to measure mRNA poly(A) tail lengths, the researchers demonstrated that MARTRE proteins protect long-tailed mRNAs from excessive deadenylation in early embryos, ensuring the efficient translation of maternal mRNAs.
Previously, several CCR4-NOT deadenylase adaptor proteins, such as BTG4 and ZFP36L2, were identified as promoting selective maternal mRNA degradation during OET in mammals. This study is the first to identify a CCR4-NOT inhibitor during OET, providing a novel perspective on maternal control mechanisms in early embryonic development. By protecting translationally active mRNAs from deadenylation, early embryos can efficiently translate limited maternal mRNAs, likely representing a universal strategy for regulating maternal gene translation during early development across species.
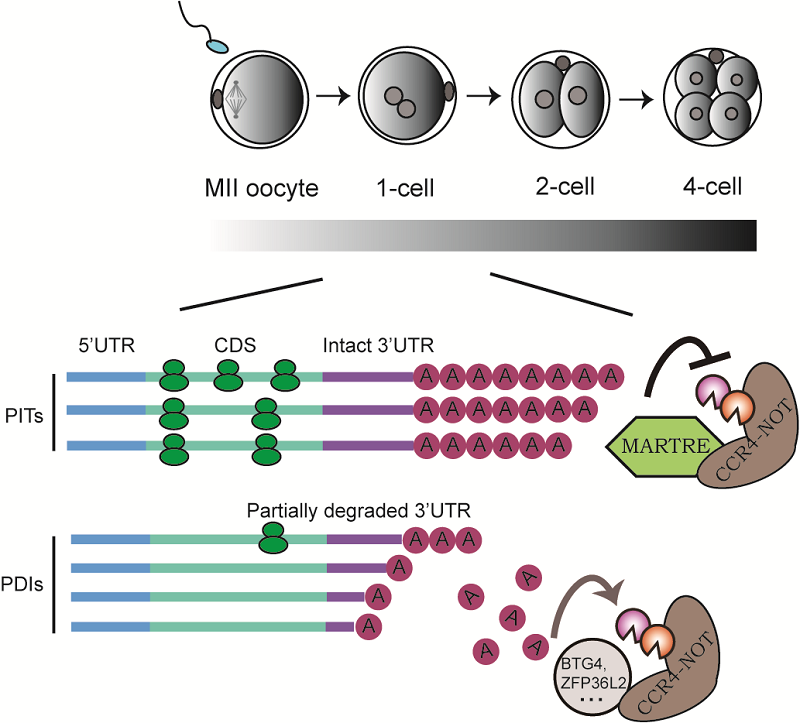
Figure: Mechanism of mRNA poly(A) tail regulation during early embryonic development
(Image by ZHU Bing's group)
Article link: https://www.nature.com/articles/s41467-024-55610-2
Contact: ZHU Bing
Institute of Biophysics, Chinese Academy of Sciences
Beijing 100101, China
E-mail: zhubing@ibp.ac.cn
(Reported by Prof. ZHU Bing's group)

