Scientists Captured High-resolution Reconstructions of Ribosome Dynamics In Situ
Ribosomal translation is crucial for life as it converts mRNA into proteins that directly carry out cellular functions. The translation process is highly complex, involving multiple stages and the precise coordination of various molecules, which makes it difficult to capture the complete translation cycle using single-particle analysis based on in vitro purification. Although studies utilizing cryo-ET have been conducted, the resolution limitations prevent a detailed representation of the dynamic multi-conformations.
In a study published in Nature Structural & Molecular Biology, a research team led by Prof. ZHANG Xinzheng from the Institute of Biophysics, Chinese Academy of Sciences, determined the large subunit (LSU) from Saccharomyces cerevisiae cell lamellae to 2.9 Å resolution using cryo-EM technology and self-developed algorithm GisSPA.
Over 20 distinct conformations were identified by 3D classification with resolutions typically higher than 4 Å. Using these conformations, they reconstruct a complete elongation cycle of eukaryotic translation with elongation factors and precisely measured the movement parameters of the small subunit (SSU) during translation.
eEF2 is the key elongation factor in the eukaryotic elongation cycle. In addition to the known extended conformations, a novel compact conformation of eEF2 was captured during the peptidyl transfer. This conformation potentially ensuring a stable environment and optimal performance for peptidyl transfer.
The researchers also discovered the binding of compact eEF2 during A-tRNA accommodation, suggesting that eEF2 associates with the ribosome much earlier than previously recognized. Additionally, a less extended eEF2 conformation was captured during the early stages of translocation. The lifecycle of eEF2 vividly demonstrates its crucial role as a 'molecular arm' in the eukaryotic elongation cycle.
Moreover, the researchers demonstrated the lifecycle of eEF3, a factor unique to certain cells like fungi, and captured a novel open conformation of eEF3. They found that eEF3 binds to the ribosome during early stage of translocation, with its conformational change coupled to head swiveling and body back-rotation of the 40S subunit.
A deeper understanding of the entire translation process helps us better uncover the intricate mechanisms of protein synthesis, and reveals its critical role in cellular functions, metabolic regulation, and the development of diseases.
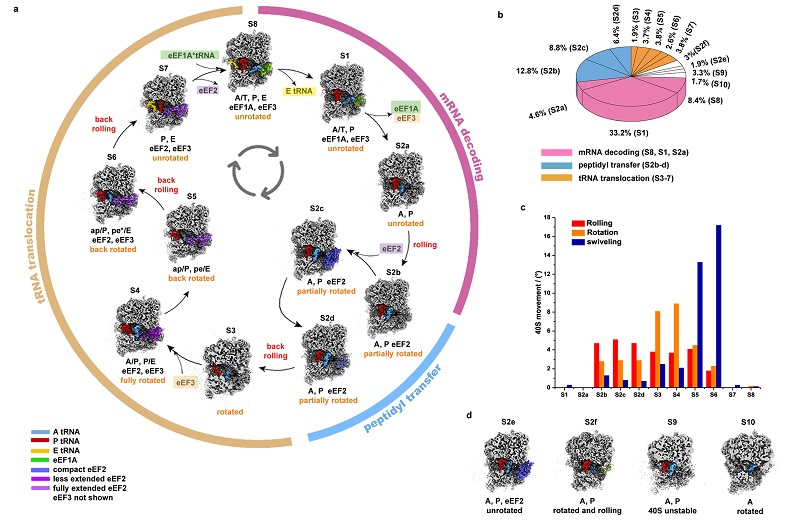
Figure 1. The major conformations of the ribosome and the elongation cycle of translation, along with the corresponding movement parameters of the small subunit
(Image by ZHANG Xinzheng's group)
Article link:https://www.nature.com/articles/s41594-024-01454-9
Contact:ZHANG Xinzheng
Institute of Biophysics, Chinese Academy of Sciences
Beijing 100101, China
Tel: 010-64888490
E-mail: xzzhang@ibp.ac.cn
(Reported by Prof. ZHANG Xinzheng's group)

