Scientists revealed the dynamic histone acetylation process by piccolo NuA4
On March 18th, 2025, the research paper "Cryo-EM structures reveal the acetylation process of piccolo NuA4" jointly completed by the research group of ZHU Ping from the Institute of Biophysics of the Chinese Academy of Sciences, and the research groups of ZHU Hongtao and LU Ying from the Institute of Physics of the Chinese Academy of Sciences was published online in the Proceedings of the National Academy of Sciences of the United States of America.
Histone acetylation is one of the most important epigenetic modifications that can induce structural changes in chromatin and regulate gene transcription. Hence, histone acetyltransferase plays a crucial role in the processes of gene regulation and DNA damage repair. Typically, most histone-modifying enzymes in epigenetic regulation, e.g., histone methyltransferases, are site-specific, that is, one modifying enzyme only modifies a specific residue in the histone tail. In contrast, many histone acetyltransferases are found to be capable of modifying multiple sites simultaneously. However, the complete process and the molecular mechanism how a histone acetyltransferase work on its multiple substrates within a nucleosome remains unclear.
NuA4 is the only essential acetyltransferase in yeast that can catalyze the acetylation of histones H2A, H2A.Z, and H4. Taking advantage of a technology named the cryo-electron microscopy single particle analysis, the research team resolved a series of structures of piccolo NuA4 (pNuA4) with various H2A.Z-containing nucleosomes, which reveal different states during the histone acetylation process by pNuA4. Furthermore, based on the solved cryo-EM structures, the researchers performed a series of single-molecule Förster resonance energy transfer (smFRET) experiments to reveal the dynamic acetylation process by pNuA4. They found pNuA4 has a much higher chance (~77%) to bind to its preferred substrate, H4, over the other substrate, H2A.Z. When pNuA4 finds its suitable substrate, amino acid residues in the N-terminal tails of H4, an amino acid residue loop (R58 - L74) in one of the component of pNuA4, Epl1, will enable it to be stably anchored on the surface of the nucleosome and initiate the acetylation process. After completing the acetylation of H4, pNuA4 will keep search for other substrates, i.e., lysine residues in H2A.Z tails, along the nucleosome surface and perform the acetylation on them. When both H4 and H2A.Z are acetylated, pNuA4 will go to a resetting state which is close to H4 and prepare for the next round of acetylation.
This study revealed a complete process of the acetylation of nucleosome by pNuA4, and clarified the specific mechanism of pNuA4 when acetylating multiple lysine substrates, and showed the dynamic sequence and structural model of pNuA4 during the acetylation of the nucleosome (Figure 1). This achievement helps researchers to understand the mechanism of gene transcription regulation by histone acetylation, and provides new insights for the treatment of related diseases.
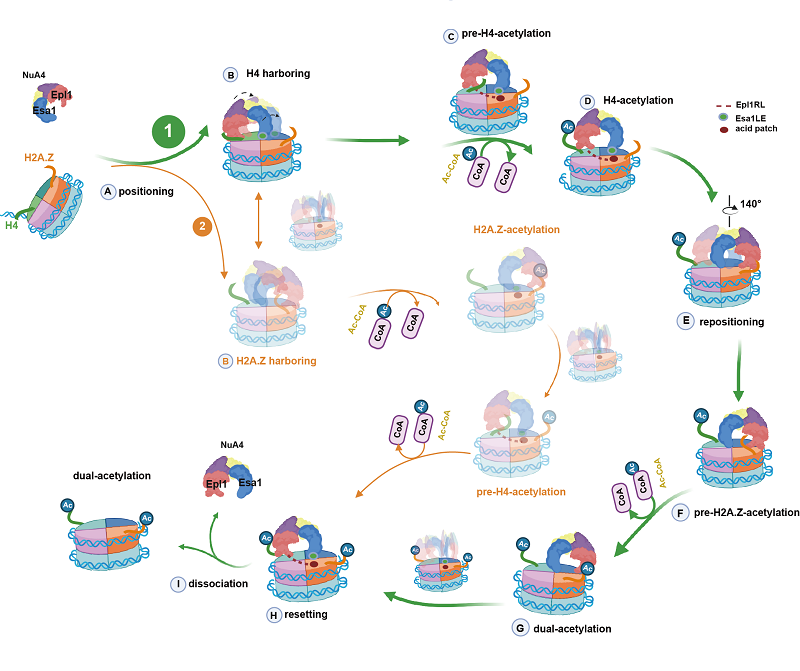
Figure 1 Schematic diagram of the process of pNuA4 acetylating different substrates on the H2A.Z-containing nucleosome
(Image by ZHU Ping 's group)
Article link: https://www.pnas.org/doi/epub/10.1073/pnas.2414490122
Contact: ZHU Ping
Institute of Biophysics, Chinese Academy of Sciences
Beijing 100101, China
E-mail: zhup@ibp.ac.cn
(Reported by Prof. ZHU Ping 's group)

