Nucleosome-Binding Protein Database Built with AI Unveils New Insights into Chromatin Regulation
The nucleosome-the fundamental unit of chromatin-plays a central role in precise gene regulation through its interactions with a wide array of proteins. Understanding the structural features and interaction mechanisms of nucleosome-binding proteins is essential for advancing epigenetics, uncovering disease mechanisms, and developing innovative therapeutic strategies. Yet, traditional experimental approaches to studying these complex interactions remain costly and time-consuming.
A research team led by Prof. ZHOU Zheng at the Institute of Biophysics of the Chinese Academy of Sciences, collaborated with Prof. XU Chunfu at the National Institute of Biological Sciences, Beijing, has leveraged the latest AlphaFold3 artificial intelligence model to systematically screen over 7,600 human nuclear proteins. This large-scale analysis produced 38,390 structural models and, for the first time, enabled the creation of a comprehensive online database and analysis platform for nucleosome-binding proteins (http://bigdata.ibp.ac.cn/ncpbindersdatabase-app, http://bigdata.ibp.ac.cn/analysis-app).
The study was published in Nucleic Acids Research on August 6, 2025.
A highlight of the study is the development of an original Strength Factor (SF) scoring system, which quantifies the strength of protein-nucleosome interactions-essentially assigning each protein a "handshake score" with the nucleosome.
This system integrates two critical measures: (1) spatial distance constraints to assess direct physical contacts, and (2) confidence metrics (PAE and pLDDT) to evaluate the reliability of predicted structures. These criteria allow researchers to pinpoint genuine nucleosome-binding proteins within massive structural datasets.
The platform itself supports multiple file formats (PDB, CIF, JSON), offering the community a versatile tool for in-depth structural analysis.
Using this framework, the team identified ARID4A and ARID4B as previously unrecognized nucleosome-binding proteins. Experiments confirmed that the HBD domains of ARID4A/4B specifically engage with the nucleosome acidic patch, and key interaction residues were mapped.
The researchers also uncovered that dimerization of RNF168, a ubiquitin E3 ligase, is essential for its stable association with nucleosomes. This discovery, validated by cryo-electron microscopy, provides critical clues to the mechanisms of DNA damage repair. Strikingly, the study further showed that AlphaFold3 can capture protein-nucleosome interaction details that are often missing from experimentally resolved structures.
This work introduces a rapid and effective computational strategy for the discovery and characterization of nucleosome-binding proteins. Beyond serving as a valuable resource for chromatin biology, the new database and analysis platform hold significant promise for advancing epigenetic research, clarifying disease pathways, and informing the development of targeted therapeutics.
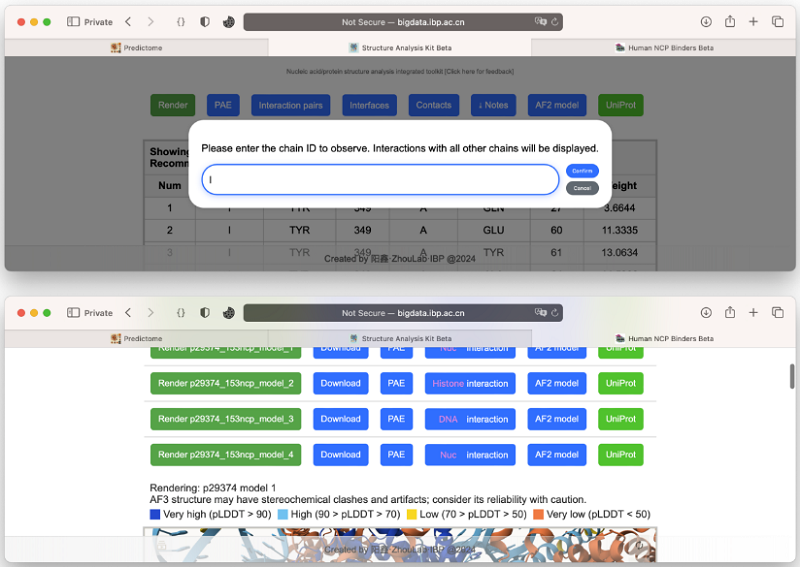
Figure 1. Online Database and Analysis Platform for Nucleosome-Binding Proteins
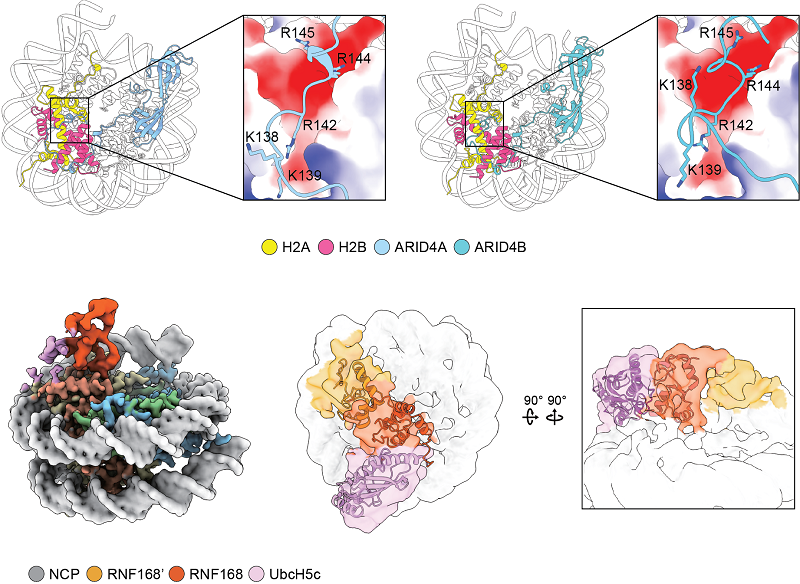
Figure 2. ARID4A/4B Recognition of the Nucleosome and RNF168 Dimerization Enhance Nucleosome Interactions
(Image by ZHOU Zheng's group)
Article link: https://academic.oup.com/nar/article/53/14/gkaf735/8223175
Contact: ZHOU Zheng
Institute of Biophysics, Chinese Academy of Sciences
Beijing 100101, China
E-mail: zhouzh@ibp.ac.cn
(Reported by Prof. ZHOU Zheng's group)

