ANGEL: A Universal Technique to Visualize, Label, and Manipulate Endogenous Proteins via Small-Peptide Knock-In
The human genome encodes over 1,000,000 proteoforms from approximately 20,000 genes, making it essential to study protein localization, expression, and interactions within their native context. Conventional overexpression of tagged proteins is often ineffective for ultrahigh-molecular-weight proteins. Moreover, it frequently leads to non-physiological expression levels that can disrupt natural protein function-especially in rhythm-sensitive proteins or those that influence organelle morphology, such as CKAP4 and RTN4.
While CRISPR-mediated endogenous tagging with fluorescent proteins preserves native regulation, large insert sizes reduce editing efficiency. Small epitope tags (e.g., ALFA) improve efficiency but lack fluorescence, posing challenges for screening monoclonal knock-in clones.
A team of researchers led by XU Pingyong at the Institute of Biophysics, Chinese Academy of Sciences (CAS) has developed ANGEL (ALFA Nb-Guided Endogenous Labeling), a novel technique that overcomes the bottleneck in high-throughput screening of cells with small peptide knock-ins. Their results were published in Nature Chemical Biology.
Through targeted mutagenesis and screening of the ALFA nanobody (NbALFA), researchers developed a modified NbALFA that functions as an antigen-stabilized fluorescent protein fusion. In the absence of the ALFA peptide, the fluorescently labeled NbALFA undergoes selective degradation. Successful insertion of the ALFA peptide into the genome triggered a substantial increase in NbALFA fluorescence intensity, selectively illuminating cells with successful ALFA-tag knock-ins.
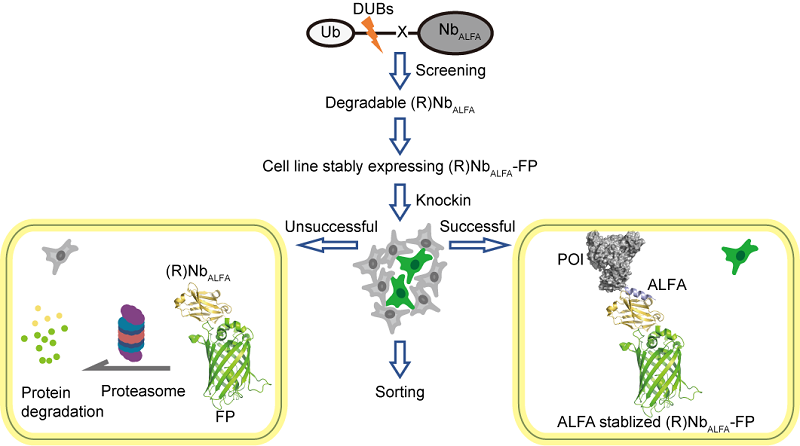
Figure. Schematic of the ANGEL process
(Image by XU Pingyong's group)
This technology integrates five key functions in native cellular environments: precise endogenous protein labeling, tandem-tag signal amplification, real-time protein dynamics tracking, targeted degradation induction, and protein interaction network analysis-offering a universal platform for live-cell research.
Article link: https://www.nature.com/articles/s41589-025-02019-7
Contact: XU Pingyong
Institute of Biophysics, Chinese Academy of Sciences
Beijing 100101, China
E-mail: pyxu@ibp.ac.cn
(Reported by Prof. XU Pingyong's group)

