Apoptosis is an important cellular proc ess that removes excess or damaged cells during organism development and adult tissue maintenance. Apoptotic cell corpses are rapidly engulfed and degraded. The failure of apoptotic cell clearance contributes to autoimmune disorders while excessive apoptosis has been associated with chronic neurodegenerative diseases. Autophagy is an intracellular degradation process, in which a double-membrane structure engulfs a portion of the cytosol and targets it for lysosomal degradation. Previous genetic studies suggested that autophagy genes are required for apoptotic cell clearance, in exposing engulfment signals on apoptotic cell surface that can be recognized by phagocytes and initiate clearance. However, the exact cellular function of autophagy genes in apoptotic corpse removal in a live organism remains elusive.
In a recent study,Professor OU Guangshuo at the Institute of Biophysics, Chinese Academy of Sciences and his colleagues used the apoptotic cell in the C. elegans Q neuroblast lineage as the model system to study the function of autophagy genes in apoptotic corpse clearance. They reported that rather than acting in apoptotic cell, the autophagy genes atg-18 and epg-5 actually function within the phagocyte to promote corpse degradation. Their observations suggested that autophagy proteins from phagocyte are recruited onto the surface of engulfed Q cell corpse and they regulate the fusion process of cell corpse with endosome and lysosome, and eventually promote the corpse clearance. These findings advanced our understanding of the contribution of autophagy genes in apoptosis.
To visualize the sequential recruitment of autophagy and other related proteins onto apoptotic cell surface, OU’s group modified a mammalian blue fluorescence protein, TagBFP, to faciliate its expression in nematodes. In combination with GFP and mCherry, they developed triple-fluorescence-protein labeling and time-lapse imaging techniques in live C. elegans.
This work was collaborated by Professor OU Guangshuo‘s group at the Institute of Biophysics, Professor WANG Xiaochen’s group at the National Institute of Biological Sciences and Dr. Ron VALE’s group at the University of California, San Francisco/ Howard Hughes Medical Institute. Dr. OU, Dr. WANG and Dr. VALE are the co-corresponding authors in this work. This work was published in the April 02, 2012 issue of Journal of Cell Biology as a research report entitled “Autophagy genes function sequentially to promote apoptotic cell corpse degradation in the engulfing cell” (http://jcb.rupress.org/content/early/2012/03/21/jcb.201111053).
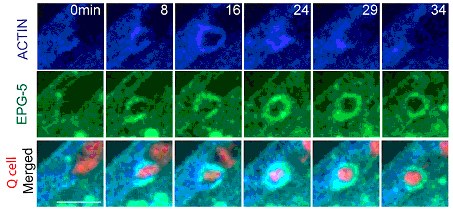
Figure: Autophagy proteins from phagocyte are recruited onto the apoptotic cell surface. (From OU Guangshuo et. al)