Genomic DNA in eukaryotic cells is packaged into a highly ordered chromatin structure. The basic repeating unit of chromatin is the nucleosome, which consists of ~146bp DNA wrapped around a histone octamer assembled from two copies of histones H2A, H2B, H3 and H4 each. Chromatin is a dynamic structure that regulates transcription, replication, repair and recombination. Posttranslational modifications of histones, including methylation, acetylation, phosphorylation and ubiquitination, play important roles in regulating chromatin structure.
In 2007, Dr. ZHANG Zhiguo and Dr. XU Rui-Ming’s labs reported that histone H3K56 acetylation is catalyzed by a unique histone acetyltransferase Rtt109 in budding yeast (Han et al., Science 2007). Unlike most other histone modifications, H3K56 is located in the N-terminal helix (αN) of H3 and is required to maintain chromatin integrity. It interacts with DNA at the entry and exit points of nucleosome. Yeast cells lacking H3K56 acetylation are highly sensitive to DNA-damaging agents and exhibit a significant increase of spontaneous chromosome breaks. Further studies show that two histone chaperons, Asf1 and Vps75 play important roles in the stimulation of Rtt109 activities (Han et al., JBC 2007a, 2007b). In particular, Asf1 is required for H3K56 acetylation in vivo, and in vitro it greatly stimulates the H3K56 acetylase activity of Rtt109. Yet, the molecular mechanism of Asf1 in regulation of Rtt109 activity is unknown.
To explore the molecular mechanism of Asf1 in H3K56 acetylation, the team of researchers lead by Dr. XU Rui-Ming at Institute of Biophysics (IBP) of Chinese Academy of Sciences (CAS) determined the crystal structure of Rtt109 from a pathogenic fungusin complex with Asf1-H3-H4 and coenzyme A (CoA). By using A. fumigatus Rtt109 (AfRtt109) which lacks the Vps75 binding region, they successfully assembled Rtt109-Asf1-H3-H4 complexin vitro and developed a simplified system for investigation of H3K56 acetylationby eliminating the complications due to Vps75 binding. The Rtt109-Asf1-H3-H4 and CoA complex structure reveals that the αN helix of H3 is unwound to facilitate Rtt109 binding to H3K56. Remarkably, although Asf1 is essential for H3K56 acetylation, Asf1 does not contact Rtt109 directly. Instead, Asf1 holds the C-terminal β-strand of H4 in a rigid conformation to facilitate optimal interaction with Rtt109. Furthermore, an interaction between Rtt109 and the centralhelix of histone H3 is also required. The structure and associated biochemical and genetic studies provided new insights into the function of histone chaperones in post-translational modifications of histones.
This first structure of a histone-modifying enzyme in complex with a multiprotein substrate complex reveals intricate contacts between Rtt109 and the Asf1-H3-H4 complex to achieve specificity. The multivalent enzyme-substrate interaction feature seen in this study may provide a glimpse into yet undisclosed intricate means of substrate recognition and regulation of enzymatic activities.
This work was published in the August 9th issue of Cell. The work was a result of collaboration between Dr. XU Rui-Ming’s group at IBP, CAS and Dr. ZHANG Zhiguo’s group at Columbia University. ZHANG Lin, a PhD student from XU Rui-Ming’s lab and SERRA-CARDONA Albert from ZHANG Zhiguo’s lab are co-first authors in this publication.
The research was supported by grants from the National Natural Science Foundation of China, including fund for Creative Research Groups, Funds for International Cooperation and Exchange (to XU Rui-Ming and ZHANG Zhiguo), Ministry of Science and Technology of China and NIH of USA. The study is further augmented by the Strategic Priority Research Program of Chinese Academy of Sciences and a Beijing Municipal Science and Technology Project grant.
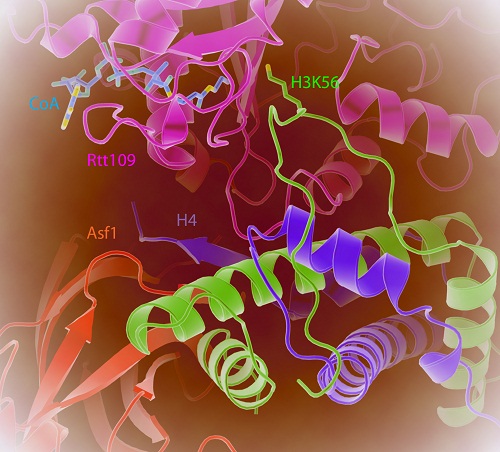
Figure. The crystal structure of Rtt109-Asf1-H3-H4-CoA complex. The N-terminal helix of histone H3 unwinds for K56 acetylation; Asf1 controls H3K56 acetylation by stabilizing the C-terminal tail of H4.
Article link: https://www.cell.com/cell/fulltext/S0092-8674(18)30899-7
Contact: XU Rui-Ming
Institute of Biophysics, Chinese Academy of Sciences
Beijing 100101, China
Phone: 86-10-64888797
Email: rmxu@ibp.ac.cn